Fig. 5.
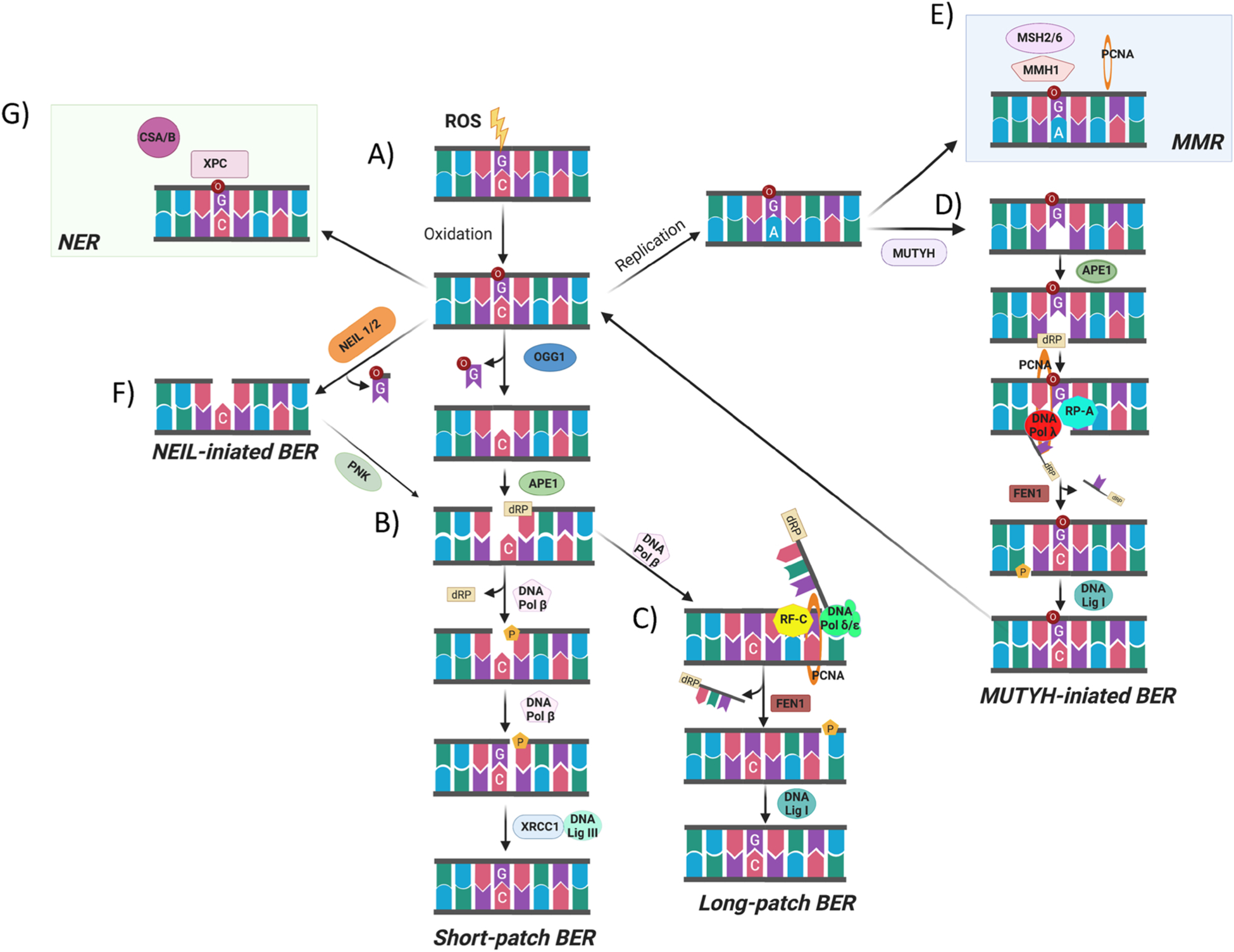
Scheme of 8-oxodG repair pathways. (A) When ROS attack DNA, this leads to the oxidation of 2′-deoxyguanosine and the formation of 8-oxodG which can pair with deoxycytidine. (B) The majority of 8-oxodG is recognized and removed by the bifunctional DNA glycosylase OGG1 through the short patch BER. Upon the removal of the oxidized nucleobase, the DNA polymerase β repairs the single-nucleotide gap by inserting a 2′-deoxyguanosine and then the DNA ligase III/XRCC1 complex mediates the final ligation step. (C) If the 5-deoxyribose phosphate (5′-dRP) terminus cannot be processed by SSB-end-processing enzymes, the DNA pol β incorporates the first nucleotide into the nick and the DNA pol δ/ε performs strand displacement synthesis creating a 5′-flap structure which is, in a PCNA-dependent manner, recognized and excised by the endonuclease activity of FEN-1. Finally, the DNA ligase I coordinates the final ligation step in long-patch BER. (D) When the 8-oxodG is not recognized by OGG1 before the S-phase, the replicative polymerase could incorporate 2′-deoxyadenosine and the resulting 8-oxodG:dA mispair may be processed by MMR components such as MSH2/6,MMH1and PCNA, to remove the mis-paired native 2′-deoxynucleoside. (E) Alternatively, the 2′-deoxyadenosine introduced during the replication could be recognized and removed by MYH mediated BER. (F) NEIL1 or NEIL2 could also initiate the 8-oxodG repair process. (G) the CSA and CSB together with XPC, as components of the TC-NER pathway, have been proposed also to repair 8-oxodG (created with biorender.com).
