Fig. 9.
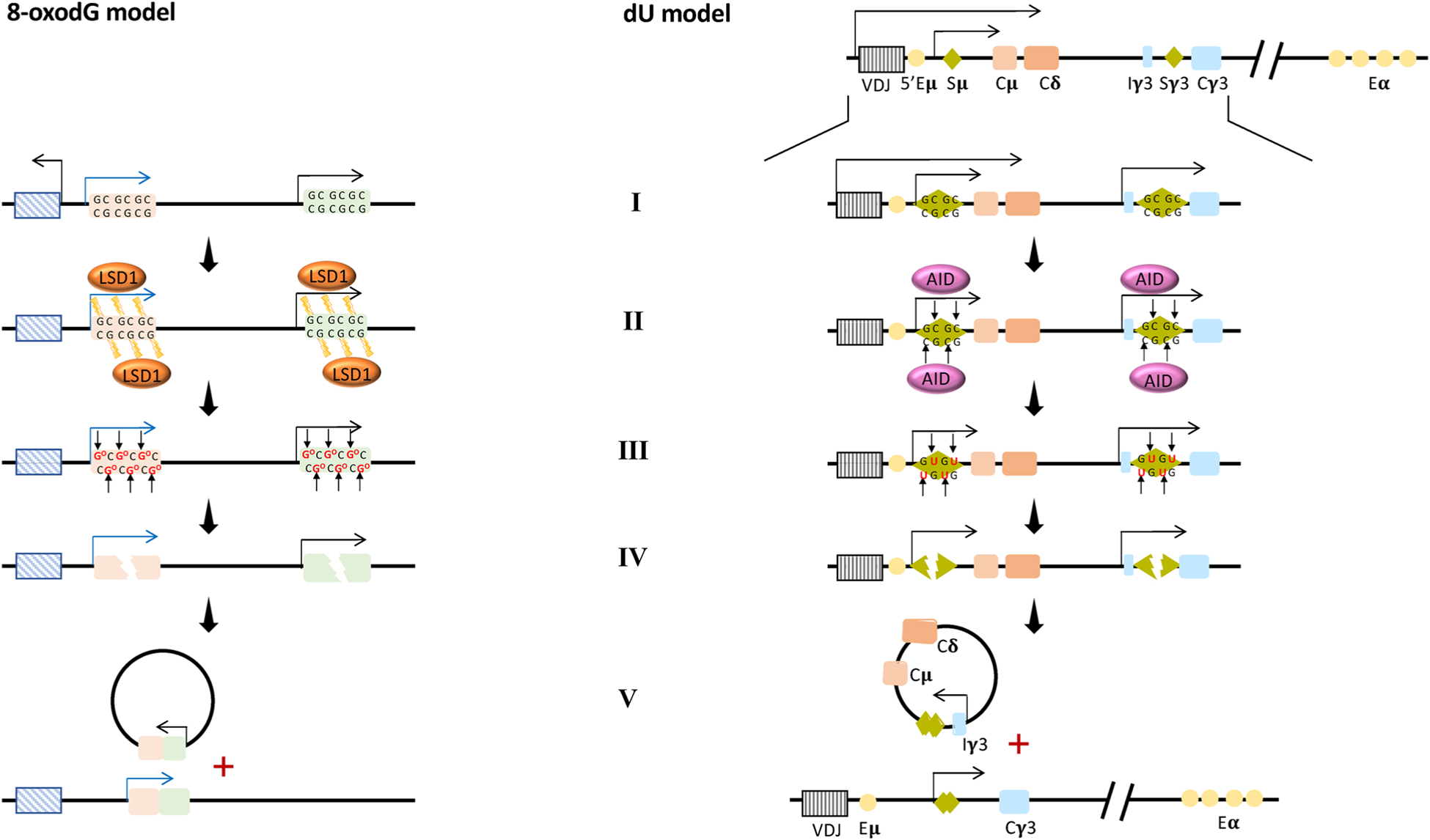
Proposed model of 8-oxodG-mediated genome instability (8-oxodG model) analagous to the AID/dU-mediated model of Class Switch Recombination (dU model).
8-oxodG model: Schematic representation of a genetic locus showing two genes subject to 8-oxodG-mediated transcription initiation. AID/dU model: Organization of the IgH locus in mice [including the antigen recognition V(D)J gene segment, the switch (S) regions, the constant (C) region exon segments, and the enhancers (E)] and associated CSR process.
(I) Specific stimuli initiate the transcription process that determines R-loops formation and RNAPII accumulation at respectively the S regions of the dU model and at oxidized promoters of the 8-oxodG model. (II–III) The recruitment of Activation-induced Cytidine Deaminase (AID) at the S regions in the dU model and the recruitment of LSD1 at the promoters of the genes loci in the 8-oxodG model (II) determine the accumulation of deoxyuridine respectively at the S regions in the dU model and of 8-oxodG at the promoters in the 8-oxodG model (III). (IV) The recruitment of specific BER enzymes: the uracil-DNA glycosylase UNG in the dU model and OGG1 in 8-oxodG model. These remove dU and 8-oxodG, respectively, generating multiple AP sites. These AP sites are processed to SSB and then to DSB by the APE1/APE2/MMR protein activities in both models. (V) Finally, two regions (each one containing one DSB) are brought together as a result of chromatin looping and ligated by the NHEJ proteins, causing a genetic translocation and deletion in both the models.
