Figure 1. Mouse ES cells provide a model system for studying the mammalian CMG helicase.
-
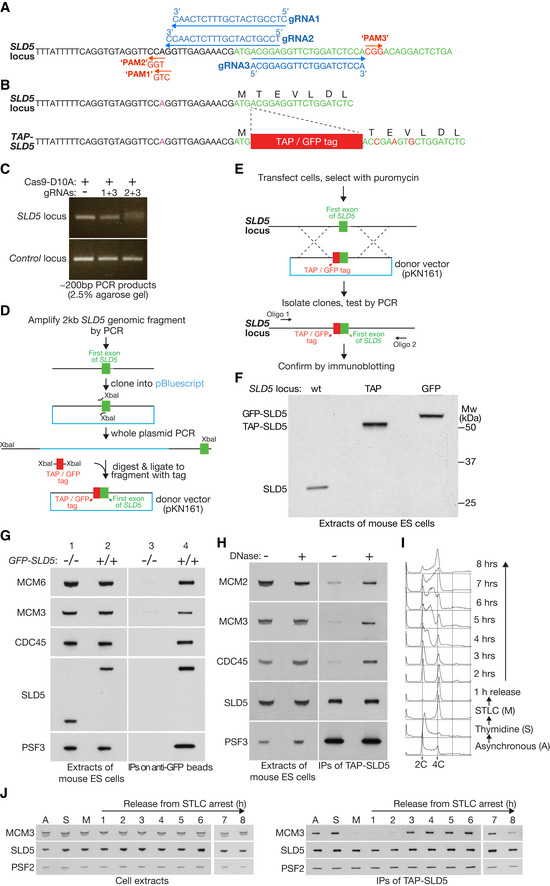
AGuide RNAs used to target the 5’ end of exon 1 of the SLD5 gene in mouse ES cells. Each of the targeted sites contains 20 nt homology to the corresponding gRNA, followed by a 3 nt “Protospacer Adjacent Motif” of PAM that has the form “NGG” and is also required for cleavage by Cas9. Note that the predicted PAM site of gRNA1 does not match the “NGG” consensus, due to a polymorphism in E14TG2a ES cells, in comparison to the reference mouse genome sequence. This polymorphism prevents cleavage by Cas9 in combination with gRNA1 (see below).
-
BThe TAP or GFP tag was inserted after the initiator methionine of SLD5. The donor DNA also contained silent mutations in the first few residues of SLD5 (indicated in red), to prevent the modified locus being cut by Cas9.
-
CPCR analysis of a pool of transfected cells after treatment with Cas9‐D10A “nickase” and the indicated combination of gRNAs from (A). The ~ 200 bp product was centred around the start of exon 1 of the SLD5 gene. Efficient cutting of both template strands by Cas9 nickase was indicated by the production of a smeary PCR product (gRNAs 2 + 3), reflecting DNA repair that mostly produced small deletions around the nicking site (e.g. see Fig EV4D below). The combination of gRNAs 1 + 3 did not produce a smeary PCR, due to a polymorphism within the PAM motif of the genomic locus (see A above).
-
DOutline of the construction of donor vectors with TAP or GFP tags flanked by 1 kb of 5’ and 3’ homology sequences.
-
EStrategy for generation of mouse ES clones with tagged SLD5 (TAP or GFP).
-
FImmunoblot indicating successful tagging of both alleles of SLD5 with TAP or GFP.
-
GAsynchronous cultures of the indicated cells were used to generate cell extracts, which were then incubated with agarose beads coupled to anti‐GFP nanobodies. The indicated factors were monitored by immunoblotting.
-
HExtracts of TAP‐SLD5 mouse ES cells were incubated plus or minus DNase for 30 min at 4°C, before immunoprecipitation of TAP‐SLD5.
-
ITAP‐SLD5 cells were synchronised as indicated (further details in Materials and Methods). DNA content was monitored throughout the experiment by flow cytometry.
-
JSamples from (C) were taken at the indicated times and used to isolate TAP‐SLD5 by immunoprecipitation on IgG beads. The indicated proteins were monitored by immunoblotting. For reasons of space, samples A, S, M and 1–6 were resolved in a separate gel to samples 7–8.
Source data are available online for this figure.
