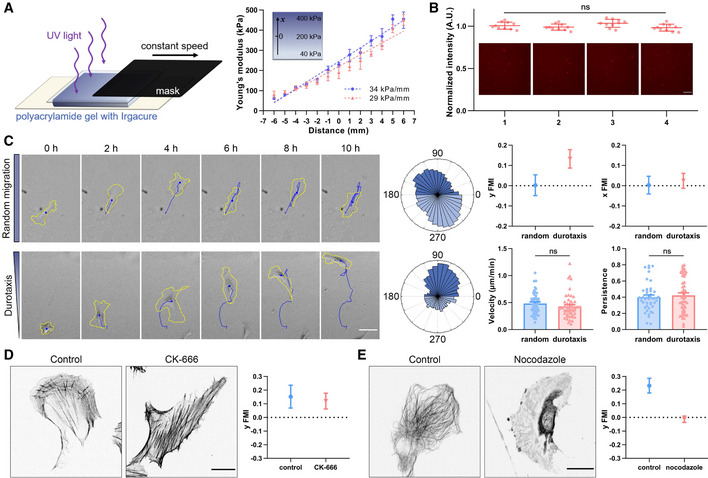
Left: Schematic diagram of manufacturing polyacrylamide gels with stiffness gradient. The gel containing photoinitiator is solidified by ultraviolet light irradiation. Mask with a constant speed is used to control the irradiation time for a hardness gradient. Right: Representative stiffness gradient of polyacrylamide gels measured by atomic force microscope was plotted (34 and 29 kPa/mm, respectively). All error bars are s.e.m. (n = 5 regions, replicates are biological).
Representative images of fluorescently labeled fibronectin at the gradient gel surface and the quantification of fluorescence intensity. 1–4 in turn represent hard to soft areas. Scale bar: 100 μm. All error bars are s.e.m. (n = 10 regions, replicates are biological), ns, not significant, by one‐way ANOVA.
Left: Representative time‐lapse images showing the migration of single hRPE cells on the surface of polyacrylamide gels with uniform stiffness (top) and stiffness gradient (bottom), referring as random migration and durotaxis migration, respectively. Yellow line is the cell outline, and blue line is the cell migration trajectory. Scale bar: 50 μm. Middle: Angular displacement of hRPE cells on the uniform stiffness gels (top) and stiffness gradient gels (bottom). Right: y/x FMI (forward migration index), velocity and persistence of hRPE cells with random migration and durotaxis. All error bars are s.e.m. (n = 50 cells, replicates are biological), ns, not significant, by Student's t‐test.
Left: Representative immunofluorescent staining of F‐actin in control and CK‐666 (100 μM, 3 h)‐treated hRPEs. Scale bar: 20 μm. Right: Y FMI of control and CK‐666 (100 μM, pre‐treated for 1 h and incubated for the whole experiment)‐treated hRPEs moving on stiffness gradient gel. All error bars are s.e.m. (n = 50 cells, replicates are biological).
Left: Representative immunofluorescent staining of α‐tubulin in control and nocodazole (8 μM, 30 min)‐treated hRPEs. Scale bar: 20 μm. Right: Y FMI of control and nocodazole (8 μM, pre‐treated for 10 min and incubated for the whole experiment)‐treated hRPEs moving on stiffness gradient gel. All error bars are s.e.m. (n = 50 cells, replicates are biological).