Figure 2. Simultaneous visualisation of histone marks and X chromosome loci in living cells.
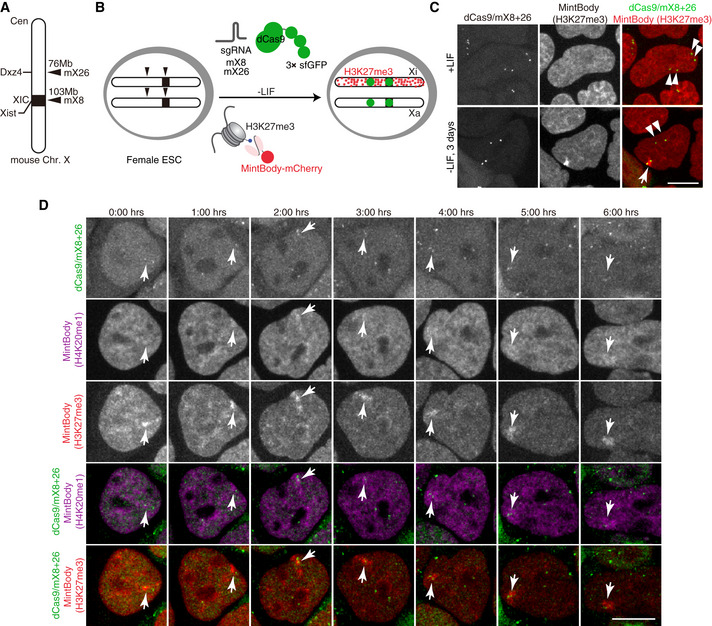
- Schematic diagram of CRISPR/dCas9‐3×sfGFP targeting loci on mouse X chromosome. sgRNA mX26 and mX8 target microsatellite repeats on Dxz4 and Xist loci, respectively.
- Experimental design: female ESCs are engineered to stably express two sgRNAs, dCas9‐3xsfGFP and a nuclear localisation signal (NLS)‐fused H3K27me3‐mintbody (mCherry). Upon ESC differentiation by LIF withdrawal H3K27me3 accumulation occurs at the inactivating X (Xi) but not at the active X (Xa). Green foci allow marking of two X‐linked loci (Dxz4 and Xist).
- Live imaging of female mESCs as in (B) in undifferentiated conditions (+LIF) or upon 3 days of LIF withdrawal. Double arrowheads mark Xa and single arrowhead marks Xi. Shown are maximum intensity projections of 11 z‐plane confocal sections. Scale bar = 10 μm.
- ESCs as in (B) were engineered to express H3K27me3‐mintbody (SNAP/JF646) and H4K20me1‐mintbody (mCherry). Cells were cultured in the absence of LIF for 2 days. Time‐lapse images for 12 z‐plane confocal stack were acquired every 1 h. Maximum intensity projection images are shown with elapse time (hh:mm). Arrowheads mark Xi. Scale bar = 10 μm.
