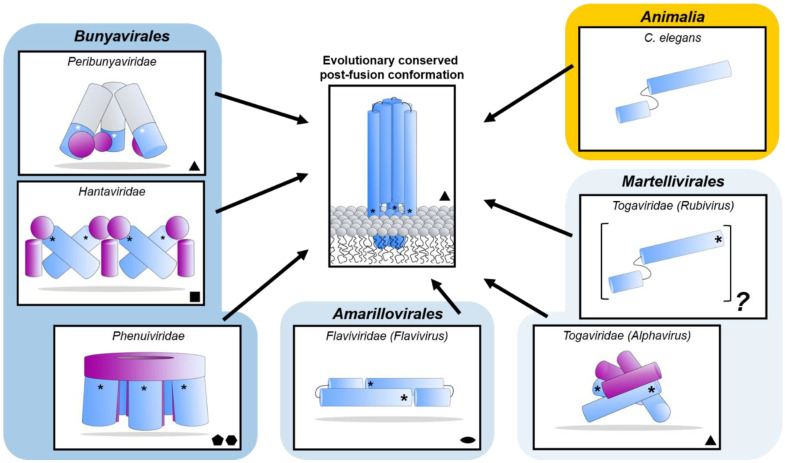
Figure 8.
Diverse class II fusion protein architectures converge on an evolutionary conserved trimeric post-fusion conformation. Schematic representation of class II fusion proteins in their pre-fusion oligomeric state. The schematic assemblies are based on crystal structures and/or cryoEM reconstructions (Peribunyaviridae: BUNV (EMD-2352 [163]) and SBV (PDB: 6H3V [153]); Hantaviridae: TULV (EMD-3364 [165]); Phenuiviridae: RVFV (EMD-4201 and PDB: 6F9F [151]); Flaviviridae: (PDB: 4UTC [189]); Togaviridae: Alphavirus eastern equine encephalitis virus (EEEV; PDB: 6MX4 [190]), Rubivirus RUBV (PDB: 4ADJ[191]); C. elegans (PDB: 4OJC [149]). The elongated structures of class II fusion proteins are shown as blue shapes (Gc for members of the Bunyavirales, E1 for Togaviridae, E for Flaviviridae and EFF-1 for the cellular C. elegans protein). Putative fusion protein stabilizing entities present on mature viral particles, are shown as purple shapes and have been hypothesized to prevent premature fusion activation (Bunyavirales: Gn, Togaviridae: E2). The E3 protein has been shown to be present in some alphavirus particles [170] but is omitted from this representation for clarity. The level of symmetry of each of the protein assemblies is indicated by symmetry symbols at the bottom right-hand corner. The approximate position of the fusion loop(s) is indicated with an asterisk (*) for each panel. In the case of peribunyaviruses, the exact location of the fusion loop (white asterisk) within the Gc protein is currently not known, but was inferred from the location of the N-terminal extensions within the tripodal EM reconstruction [153,163] and the C-terminal positioning of Gc transmembrane domains. Note that, although C. elegans EFF-1 (epithelial fusion failure 1) protein presents a class II fusogen architecture, it does not contain a fusion loop. Fusion is believed to be initiated by trimerization of the plasma membrane anchored EFF-1 ectodomains protruding in the extracellular space [149]. The grey region of the column shown for Peribunyaviridae represents the N-terminal extension of the Gc fusion protein, which has not been observed in other bunyavirus glycoproteins. The pre-fusion oligomeric state of EFF-1 has been observed to be monomeric on the plasma membrane [192]. The pre-fusion oligomeric state of rubella virus E1 on the virus membrane is currently unknown and therefore represented as a protomer of an unknown oligomeric assembly. The fusion proteins of alpha- (e.g., Semliki Forest virus (SFV), chikungunya virus (CHIKV)) and flaviviruses (e.g., dengue virus (DENV), zika virus (ZIKV)) are structurally related despite a lack of detectable sequence conservation and are therefore positioned next to each other in the diagram. Similarly, phenuivirus Gc has been shown to be genetically more closely related to the fusion envelope (E) proteins of flaviviruses than to those of other genera in its own order [152]. These proteins are placed next to each other to represent this predicted relationship. The box depicting the cellular EFF-1 protein is colored in yellow as to oppose the boxes in different shades of blue which all contain viral fusion proteins.