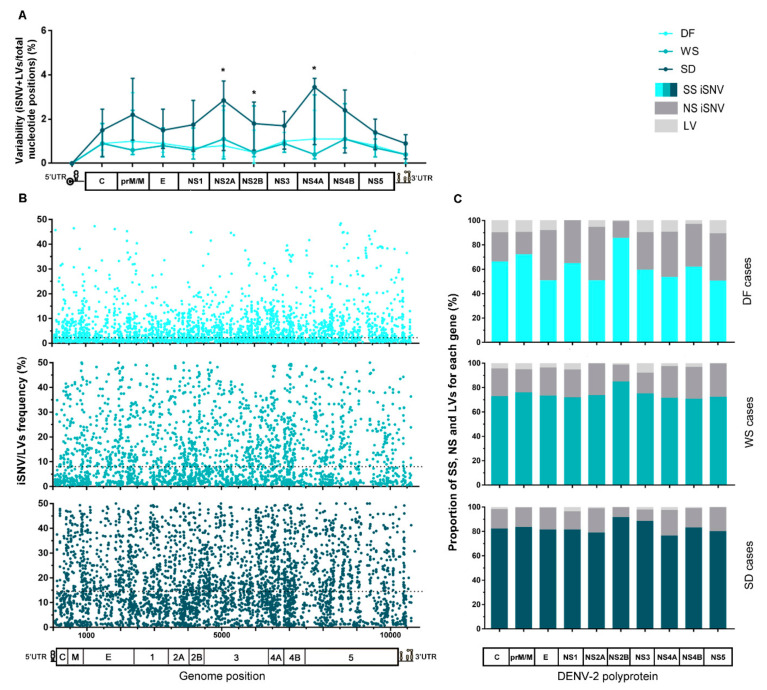
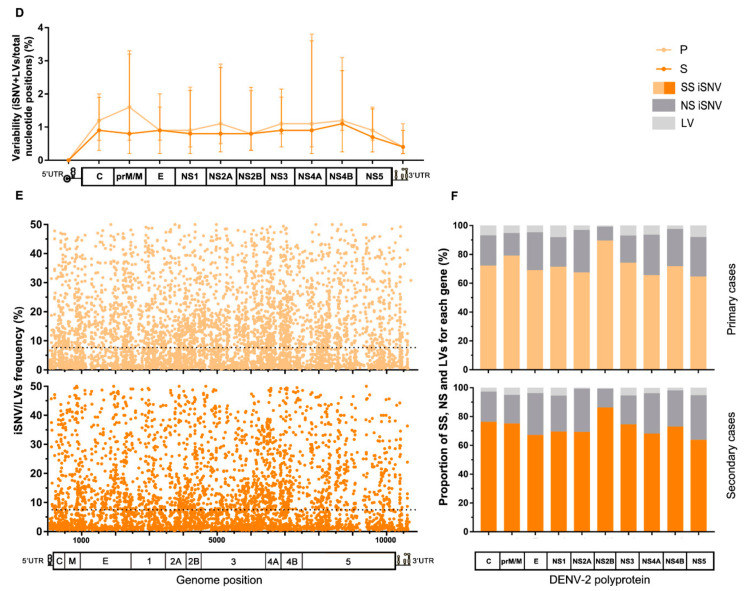
Figure 2.
Characteristics of the intrahost viral population of cases grouped by clinical category or immune response. (A) Variability along the viral genome according to the patients’ clinical outcome. The total number of iSNVs located in each gene along the viral genome was normalized by each gene’s length (total nucleotide positions) and the median value for each group plotted in the graph, with error bars representing the interquartile ranges. (*) Statistically-supported differences among clinical groups. (B) iSNV/LVs frequency distribution along the viral genome according to the clinical category. The dotted line represents the median frequency among all iSNV/LVs found within each group. (C) Percentage of synonymous, non-synonymous iSNVs, and LVs for each gene. Clinical categories are represented separately. The total amount of each variant class per gene was summed and then normalized by group size. (D–F) The same analysis was performed for cases grouped by patients’ immune response. Genome/polyprotein schemes in graphs A, C, D, and F are not scaled to genes real size, as are the schemes in graphs B and E. DF: dengue fever cases, WS: dengue with warning signs, SD: severe dengue cases, P: primary cases, S: secondary cases, SS: synonymous variants, NS: Non-synonymous variants, and LV: insertion/deletion variants.