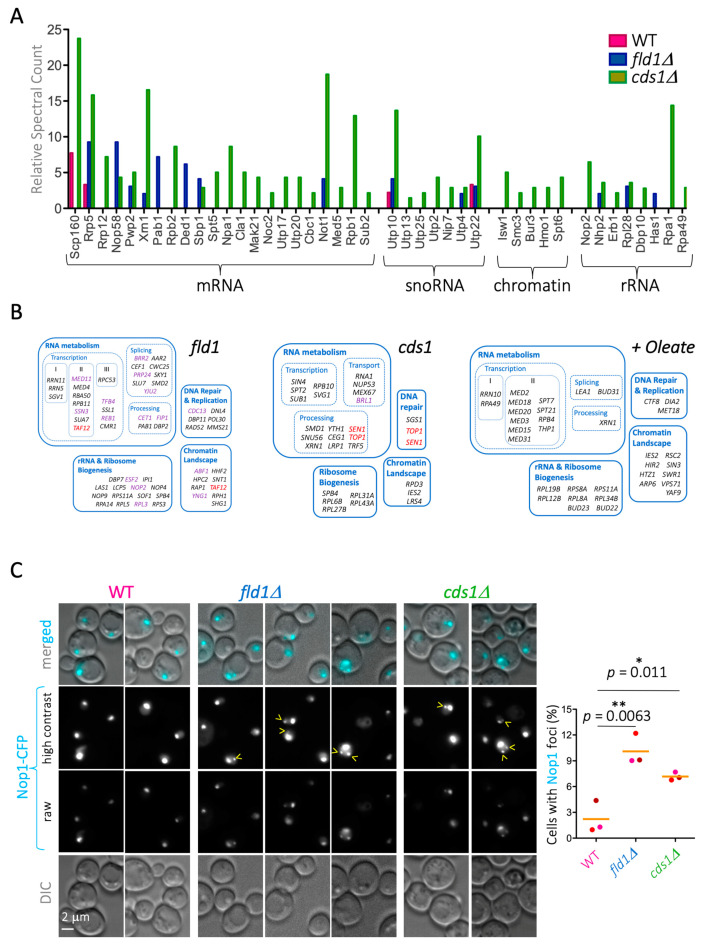
Figure 7.
LD alterations predispose to genome instability. (A) Graphical representation of the relative presence of karyopherin cargoes found on the LD of WT, fld1Δ, and cds1Δ cells purified from saturated Saccharomyces cerevisiae cultures (original data reported in [31]). The hits are grouped by biological role. (B) The 257 genetic interactors of the fld1 mutation (left) and the 232 interactors of the cds1 mutation (center) were retrieved from the Saccharomyces Genome Database, while the 134 oleate-sensitive interactors (right) were retrieved from [32]. The nuclear hits were selected, and further functional classification was performed with DAVID [33], and these are shown as individual sets. Genes in violet indicate positive genetic interactions, while the rest are negative ones. Genes in red are included twice because the protein can be ascribed to two functional categories. (C) left: Images of WT, fld1Δ, and cds1Δ cells that were transformed with a plasmid expressing the nucleolar marker Nop1 tagged with CFP. Asynchronous cultures were used for live fluorescence microscopy. Two or three examples are shown for each strain. Nucleoli are visible in the raw and high-contrast images. Nop1 foci, indicative of fragmented nucleoli, are indicated by yellow arrowheads. right: Quantification of the percentage of cells in the population with Nop1 foci. Dots represent three independent experiments. Similarly colored dots belong to the same experiment. The orange bar is the mean of those three experiments. At least 300 cells were counted per condition and experiment. The p values correspond to paired t-tests assessing the significance of the differences of the means.