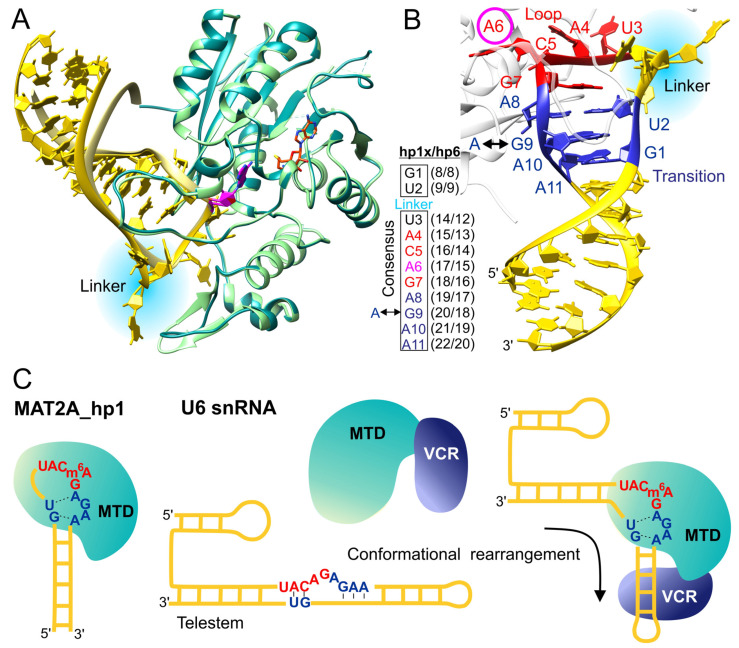
Figure 2.
Interaction between METTL16 and RNA methylation substrates. (A) Superposition of two crystal structures of METTL 16-MTD/RNA complex, PDB ID: 6du4 (METTL16, turquoise; hp1x, gold), 6du5 (METTL16, light green; hp6, light yellow); the acceptor adenosine is shown in magenta; SAH is superimposed from the structure 6b92 and shown as sticks (orange). The linker differentiating the loop-transition region of hp1x and hp6 is highlighted by light blue. (B) The hp1x of METTL16-MTD/RNA complex (6du4) is shown in atoms-ribbon representation. The 3′ part of the loop and the transition region are marked by red and blue colors, respectively. The acceptor adenosine is manifested by the magenta circle. The linker between the transition region and the loop is highlighted by light blue. METTL16 is shown by a transparent white ribbon. The conserved sequence of the loop-transition structure is numbered 1–11, with 6 (magenta) representing the acceptor adenosine. Schematic representation of loop-transition region (inset) shows conserved residues (1–11, box) and corresponding nucleotides in hp1x and hp6 models. The arrow indicates the G/A substitution in hp5. (C) A schematic diagram of the interaction between METTL16 and its RNA substrates ([36], figure modified): MAT2A mRNA and U6 snRNA. The loop and transition regions are marked as in panel B. The VCRs of METTL16 associate with U6 and rearrange the substrate to obtain the conformation optimal for methylation.