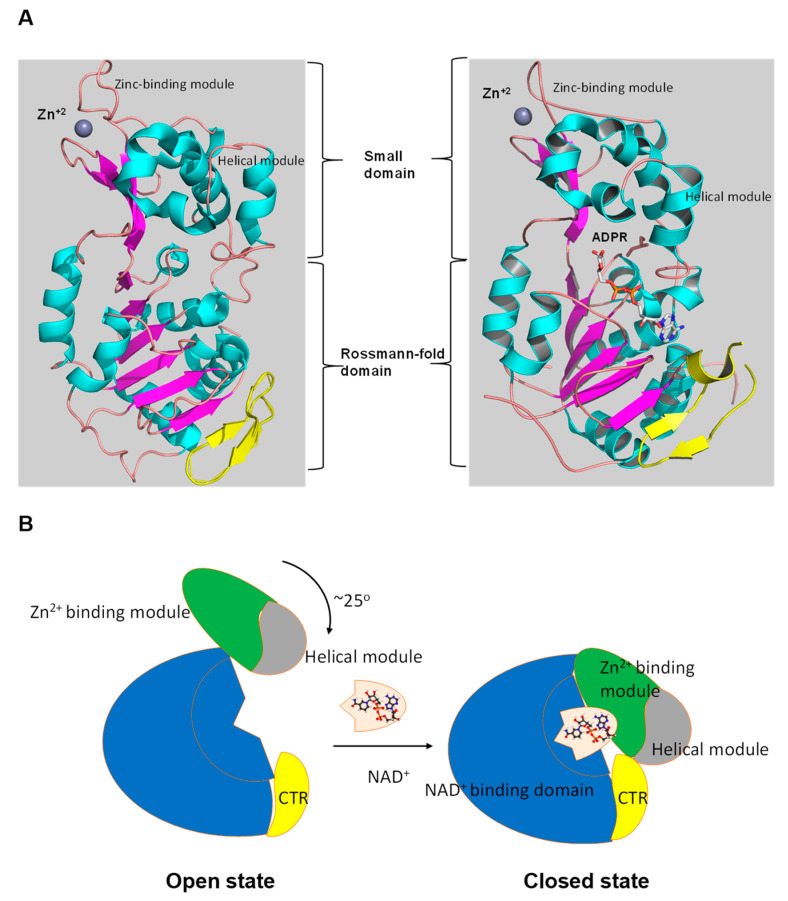
Figure 3.
The tertiary structure of SIRT1. (A) The structure in open (left) conformation (SIRT1•CTR complex, PDB code 4IF6) and in closed (right) conformation (SIRT1•CTR•ADPR•Substrate complex, PDB code 4KXQ). The amino acid sequence of SIRT1 is organized into two independent domain. A cavity that is created between the two domains forms the active site. The C-terminal regulatory segment (CTR) is shown in yellow. The CTR binds at the lower edge of the larger NAD+-binding domain, complementing the central parallel β sheet of its Rossmann fold. The figure was created using the program PyMOL (www.pymol.org). (B) Cartoon model of the conformational changes of SIRT1 upon substrate and NAD+ analogue binding. The smaller domain undergoes a rotation with respect to the large domain. A cartoon representation of the apo SIRT1·CTR heterodimer (open state) and the SIRT1·CTR·NAD+·Substrate complex (closed state). Comparison of the open and closed structures revealed that the larger NAD+-binding domain does not undergo any major structural changes. The smaller domain rotates about 25°. The small domain (Zn2+-binding module and the helical module) rotates as a rigid body with only minor changes to the backbone and sidechain conformations. Adapted from Ref. [62].