Abstract
Faithful propagation of life requires coordination of DNA replication and segregation with cell growth and division. In bacteria, this results in cell size homeostasis and periodicity in replication and division. The situation is perturbed under stress such as DNA damage, which induces filamentation as cell cycle progression is blocked to allow for repair. Mechanisms that release this morphological state for reentry into wild-type growth are unclear. Here we show that damage-induced Escherichia coli filaments divide asymmetrically, producing short daughter cells that tend to be devoid of damage and have wild-type size and growth dynamics. The Min-system primarily determines division site location in the filament, with additional regulation of division completion by chromosome segregation. Collectively, we propose that coordination between chromosome (and specifically terminus) segregation and cell division may result in asymmetric division in damage-induced filaments and facilitate recovery from a stressed state.
INTRODUCTION
For successful cell division to occur, accurate DNA duplication and segregation must be completed. In bacteria, chromosome replication initiates bidirectionally from an “origin” and finishes opposite to this position, at the “terminus” (Kleckner et al., 2014, 2018; Reyes-Lamothe and Sherratt, 2019). Several factors ensure that cells divide only upon completion of this process by regulating the multiprotein division machinery called the “divisome” (Galli and Gerdes, 2012; Männik et al., 2016; Dewachter et al., 2018; Kleckner et al., 2018). For example, Escherichia coli encode negative regulators of the tubulin homologue FtsZ that is required to initiate the assembly of the divisome at the division plane. Nucleoid occlusion by SlmA prevents the formation of the FtsZ-ring at locations where chromosomal DNA is present and MinCDE oscillations direct the position of the Z-ring near midcell (Bernhardt and de Boer, 2005; Tonthat et al., 2011; Tsang and Bernhardt, 2015). Recent studies have also suggested coordination of division with the terminus via proteins such as MatP and ZapAB that act as a bridge between the DNA as well as the divisome (Mercier et al., 2008; Espéli et al., 2012; Männik et al., 2016). Together, in unperturbed laboratory conditions, this results in daughter cells that replicate and divide in a periodic manner and that do not show much deviation in birth and division cell sizes (Donachie, 1968; Campos et al., 2014; Taheri-Araghi et al., 2015; Harris and Theriot, 2016; Wallden et al., 2016; Micali et al., 2018; Si et al., 2019). Such size maintenance has been described in other bacteria as well as eukaryotic systems (Soifer et al., 2016; Chandler-Brown et al., 2017; Lambert et al., 2018).
Recent investigations in bacteria have proposed that cells follow an “adder”-based principle, where length added between divisions is independent of the birth length of the cell. Size homeostasis is governed either via regulation at the stage of replication initiation or cell division or both (Campos et al., 2014; Taheri-Araghi et al., 2015; Wallden et al., 2016; Si et al., 2019). In addition, some studies have also proposed a role for cell shape or concurrency between processes of replication and division in size control (Harris and Theriot, 2018; Micali et al., 2018). This homeostatic state is perturbed under conditions of stress, a situation that can be often faced by bacterial cells in their environment (Justice et al., 2008; Horvath et al., 2011; Yang et al., 2016; Heinrich et al., 2019). For example, under DNA damage, a cell cycle checkpoint blocks cell division until damage has been repaired. The bacterial SOS response is activated upon the binding of RecA to single-stranded DNA that is exposed as a consequence of DNA damage (Mukherjee et al., 1998; Kreuzer, 2013; Jonas, 2014). As part of this response, a cell division inhibitor (such as SulA in E. coli) blocks polymerization of FtsZ, resulting in cellular elongation or filamentation (Mukherjee et al., 1998; Kreuzer, 2013). Cell division inhibition during damage is a conserved process, even though the effectors may vary across bacteria. SOS-induced division inhibition is carried out by SidA in Caulobacter and YneA in Bacillus (Mukherjee et al., 1998; Mo and Burkholder, 2010; Modell et al., 2011; Jonas, 2014). SOS-independent DNA damage-induced division inhibitors have also been identified, suggesting that this is an important step in DNA repair (Modell et al., 2014). Along with blocking division, chromosome cohesion is also initiated in these filaments. It is thought that this can aid recombination-based repair (Odsbu and Skarstad, 2014; Vickridge et al., 2017).
Cells have also been shown to change shape and size under other forms of stress including host environments, heat shock, and osmotic fluctuations (Justice et al., 2008; Bos et al., 2015; Kysela et al., 2016; Yang et al., 2016; Caccamo and Brun, 2018; Wehrens et al., 2018; Heinrich et al., 2019). Together, this highlights the plasticity with which bacteria such as E. coli sample a range of cell sizes including filamentous and nonfilamentous cell lengths. The process by which cell division is regulated to result in elongation under DNA damage has been well characterized (Adler and Hardigree, 1965; Kantor and Deering, 1966; Suzuki et al., 1967; Mukherjee et al., 1998; Mo and Burkholder, 2010; Modell et al., 2011, 2014; Jonas, 2014). Indeed, recent cell biological studies have also characterized division restoration dynamics of non-DNA damage-induced filaments (such as SulA overexpression or heat shock; Wehrens et al., 2018). However, unlike these stresses, DNA damage specifically perturbs chromosome organization and segregation, with replication initiation continuing despite damage to DNA (Rudolph et al., 2007; Lloyd and Rudolph, 2016). The impact of such a damage-induced state on reinitiation of wild-type-like division after repair is mechanistically less characterized.
In this study, we probe the mechanism by which filamentous E. coli reinitiate chromosome segregation and cell division after DNA repair. We use single-cell, time-resolved fluorescence microscopy to follow the kinetics of division restoration after cells face a pulse of DNA damage and observe that filamentous cells tend to divide asymmetrically, generating short daughters of wild-type size and growth dynamics. We further observe that division restoration is controlled by two steps: determining the location and timing of division. This process, regulated by a combination of MinCDE oscillations and chromosome (specifically terminus) segregation, is accompanied by asymmetric partitioning of repaired chromosomes, resulting in the production of daughter cells of wild-type size and devoid of DNA damage, thus likely facilitating recovery from a stressed state.
RESULTS
Asymmetric cell division in DNA damage-induced filaments
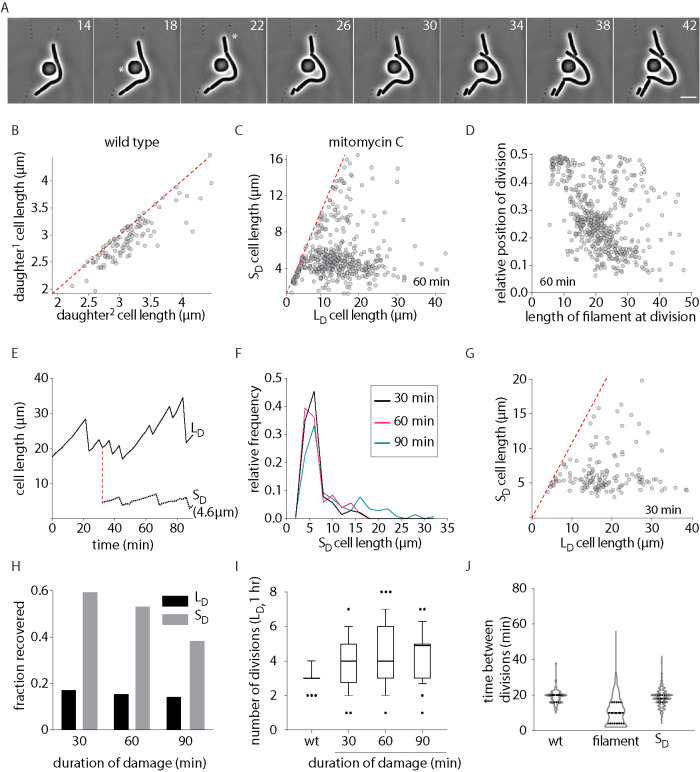
To understand how damage-induced filamentous E. coli reinitiate cell division and wild-type growth after DNA damage, we followed division restoration in cells after treatment with a subinhibitory dose of the DNA damaging agent mitomycin-C via time-lapse imaging (1 μg/ml; Dapa et al., 2017; Figure 1A). While unperturbed wild-type cells divided near midcell (Figure 1B), we found that a significant proportion of cells deviated from this division pattern as they increased in length (Figure 1, C and D). Damage-treated cells close to wild-type length (5–10 μm) divided in the middle resulting in the production of two daughter cells of similar sizes. In contrast, filamentous cells divided asymmetrically to produce a short “daughter” cell (SD) and a long cell that continued to filament (LD; Figure 1, C and E). The probability of a cell to undergo asymmetric division increased with increasing cell length with 85% of cells dividing asymmetrically at lengths >12 μm. SD production via asymmetric division was observed independent of growth media (Supplemental Figure S1A). In addition, as seen in the case of division-inhibited E. coli filaments (Wehrens et al., 2018), cephalexin-treated cells (where division is inhibited, without perturbing genome integrity; Rolinson, 1980; Chung et al., 2009) also tended to have increased non-midcell (asymmetric) divisions with increasing filament lengths (Supplemental Figure S1, B and C). Varying durations of damage exposure (30, 60, or 90 min) resulted in different degrees of filamentation. However, in each case, the SD size distribution for these treatments was comparable. Additionally, >81% of SD’s were close to wild-type size distribution (Figure 1, F and G and Supplemental Figure S1, D–G). The frequency of non-midcell division was similar between filaments of comparable lengths (12–40 μm). However, the distribution of division site locations varied with varying cell length bins (discussed further in Figure 3 and the associated section, later in the article).
FIGURE 1:
Asymmetric division in DNA damage-induced filaments during recovery. (A) Representative time-lapse montage of filamentous cells during recovery. White asterisks indicate divisions occurring toward a cell pole. Scale bar = 5 µm; time in minutes here and in all other images. Images were taken every 2 min. (B) Cell length of two daughter cells generated from a single division in wild-type conditions. Each gray dot represents a single division event (n = 157). The red line plots the expected values if all cells were dividing at their midpoint. (C) Cell length of long daughter (LD) and short daughter (SD) generated from a DNA damage-induced filament during recovery. Cells are treated with mitomycin-C (MMC) for 60 min. Each gray dot represents a single division event (n = 531). The red line plots the expected values if all cells were dividing at their midpoint. (D) Location of division is plotted as a function of cell length in filamentous E. coli during recovery from DNA damage treatment (60 min; n = 531). (E) Cell length of a long daughter (LD) and short daughter (SD) is tracked over time during damage recovery. Decrease in cell length is indicative of division. (F) Distribution of SD lengths generated from filaments between 12 and 40 µm long after 30, 60, and 90 min of MMC treatment (n = 142 [30 min], 363 [60 min], 96 [90 min]). (G) As C for cells treated with MMC for 30 min (n = 151). (H) Fate of SD and LD during recovery. Cell is classified as recovered if it undergoes midcell division and produces a daughter of wild-type size and filamentous if it continues to filament after division (n = 116 [30 min, LD], 98 [60 min, LD], 106 [90 min, LD], 150 [30 min, SD], 264 [60 min, SD], 150 [90 min, SD]). (I) Number of divisions per cell in 1 h for all durations of damage treatment. As a control, the number of divisions wild-type cells undergo is also shown (n = 50 [wt], 150 [filaments]). (J) Distribution of time between divisions for wild-type (no damage control), damage-induced filament, and SD during recovery from MMC (n = 148 [wt], 611 [filament], 468 [SD]).
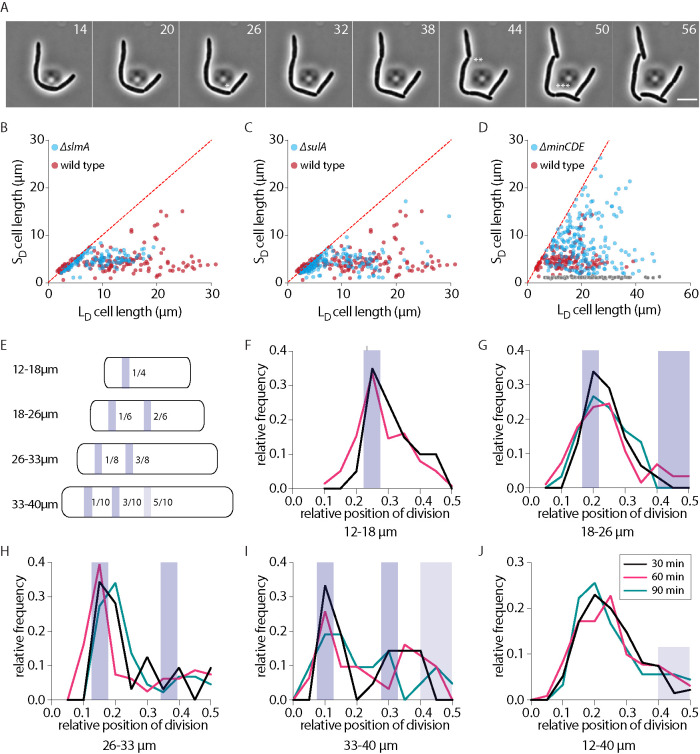
FIGURE 3:
Role of Min-system in division positioning. (A) Representative time-lapse montage of division in wild-type cells during damage recovery. (B–D) Cell length of long daughter (LD) and short daughter (SD) generated from a DNA damage-induced filament during recovery for slmA (n = 144), sulA (n = 246), and minCDE (n = 186) backgrounds, respectively (blue dots; minicells are shown in gray). As a reference, lengths for wild type (n = 137) during recovery are shown in red. The red line plots the expected values if all cells were dividing at their midpoint. (E) Schematic representation of the Min-driven division site rule (figure adapted from Wehrens et al., 2018). Location of division for various filament length bins is shown with the blue band. Precise location of division (relative to cell length) is depicted inside each cell. (F) Distribution of relative position of division for filaments between 12 and 18 µm for 30 or 60 min of damage treatment. Data are not shown for 90 min of treatment as the number of filaments in this length range in 90 min treatment is low. Location of division as determined by the Min-rule is shown as a shaded bar. (G–I) As F for filaments between 18 and 26, 26 and 33, and 33 and 40 µm, respectively. (J) As F for all filaments between 12 and 40 µm. Location of potential midcell division is shown with the shaded bar (n [all] = 135 [30 min], 453 [60 min], 95 [90 min]).
To characterize the recovery process further in DNA damage-induced filaments, we followed the fate of the filament (LD) and the SD over time. We observed that filamentous cells underwent multiple divisions in a 1 h time period, generating daughter cells (SD) of wild-type size at each division (Figure 1I). In 16 ± 2% cases, the filament itself was restored to wild-type size during the course of imaging. In the same time, wild-type cells undergo three divisions on average, suggesting that more daughter cells are produced from a filament than a cell of the same size as wild type during recovery. In contrast to the filaments, daughter cells of wild-type size displayed growth and division dynamics similar to nondamage conditions (Figure 1, H and J). As an example, in Figure 1E, we followed the fate of an SD (4.6 μm) generated from a filament and found that the daughter cell reinitiated wild-type growth dynamics soon after division. Time taken between divisions for SD was close to the distribution seen for wild type, in contrast to interdivision times observed for the nonrecovered, filamentous LD (Figure 1J). Consistently, the DNA damage marker, RecA, formed multiple foci in LD at the time of division, while SD had RecA localization as seen for wild-type cells (Lesterlin et al., 2014; Rajendram et al., 2015; Vickridge et al., 2017) or elongated cells with no DNA damage (cephalexin treated; Supplemental Figure S1, K and J). Our observations are in line with the idea of the reversible nature of damage-induced filamentation (Rudolph et al., 2007, 2010; Vickridge et al., 2017) and suggest that filamentous cells can restore cell division and recover from a stressed state. Consistently, we found that a viable cell count as well as cell length distribution was restored to close to that of wild type in the three treatment regimens within 120 min after recovery (Supplemental Figure S1, L and M). Taken together, our results are complementary to previous studies reporting non-midcell division events in filamentous E. coli (Begg and Doanachie, 1977; Taschner et al., 1988; Rudolph et al., 2009; Wehrens et al., 2018). We thus characterized the factors contributing to SD production in the case of damage-induced filaments.
Division dynamics of damage-induced filaments
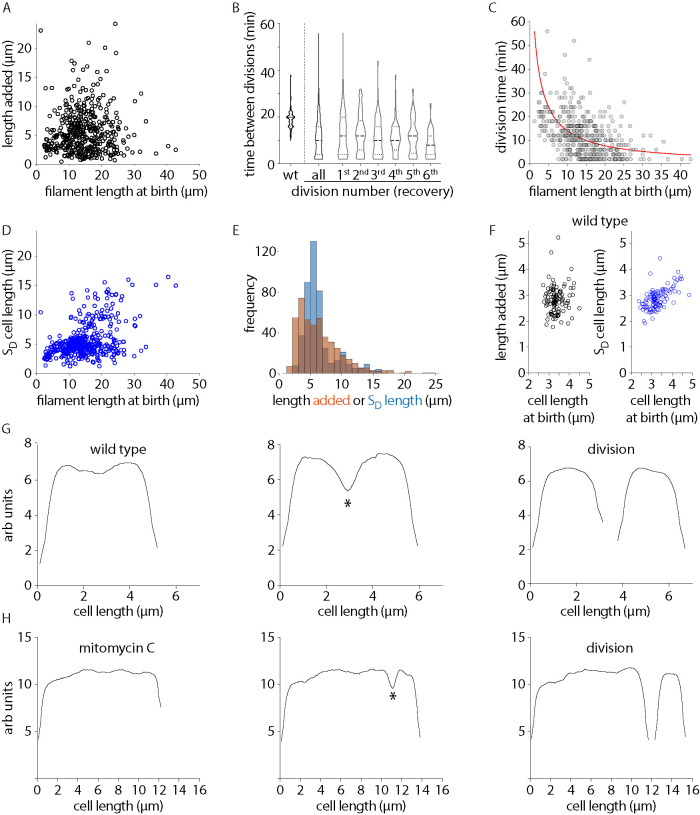
In the case of wild-type steady-state population, where cells divide at midcell, the adder principle contributes to size homeostasis. Under adder, length added between divisions is thought to be independent of the length of the cell at birth (Campos et al., 2014; Taheri-Araghi et al., 2015; Wallden et al., 2016; Si et al., 2019). To understand what determines the distribution of short daughter sizes in the case of asymmetrically dividing damage-induced filaments, we further analyzed the growth and division dynamics of these filaments (details of cell division analysis are outlined in Figure 2, G and H, Supplemental Figure S2, D and E, and Materials and Methods). In damage-induced filaments, we found that length added between divisions did not correlate with length of the cell at birth. There were several instances of consecutive divisions without detectable elongation in between each division (Figure 2A). We also measured time between consecutive division events. While wild-type cells divided approximately every 20 min, filamentous cells displayed a large distribution of times. However, on average, longer cells tended to have shorter interdivision times, consistent with the observation that filamentous cells underwent an increased number of divisions when compared with wild type (Figure 2, B and C). In addition, we found that 67 ± 4% of divisions took place at the opposite pole of the previous division. This suggests that division did not preferentially occur only toward a single pole (old or new), with several instances of division events taking place in an alternating manner.
FIGURE 2:
Division dynamics of damage-induced filaments. (A) Length added to a cell between divisions as a function of the birth length of the cell for MMC-treated cells (n = 452), p value 0.0293, 95% CI [−0.0631 0.1212], Pearson correlation value via MATLAB corrcoef function. (B) Time between divisions for wild-type cells or damage-recovery filaments for all divisions and for cells between first and second, second and third, and so on until the sixth division in filaments during recovery is plotted (n = 148 [wt], 410 [all filaments]). (C) Time between divisions as a function of cell length at birth for MMC-treated cells. Red line shows the relation that would be necessary for the system to be an adder (Eq. 3 in supplementary results), given that cells are growing exponentially with the rates given in Supplemental Figure S2A (n = 458). (D) SD cell length as a function of the birth length of the cell for MMC-treated cells (n = 457, p value 0.4650, 95%CI [0.3894 0.5343], Pearson correlation value via MATLAB corrcoef function). (E) Distribution of length added and SD cell length at each division for MMC-treated cells (distributions are significantly different, two-sample Kolmogorov-Smirnov test, p value 1.0721 × 10−8, n = 452). (F) As A and D for wild-type cells (length added, n = 107, p value 0.0647, 95%CI [−0.1268 0.2514]; length removed, n = 107, p value 0.6057, CI [0.4698 0.7135], Pearson correlation value via MATLAB corrcoef function). (G) Phase profile for a wild-type cell before and after division is plotted. Constriction is marked with an * and divisions are identified during segmentation. (H) As G for a MMC-treated filament.
After damage, filament length increased at a characteristic rate, as seen in wild-type conditions (Supplemental Figure S2, A and B), with some fluctuation which probably arises because of stochasticity in damage and/repair. As stated above and expected for the adder principle seen in wild-type conditions, we found that length added between divisions had no correlation with birth length of filaments as well (Figure 2A). However, we noticed the following deviations from wild-type growth and division patterns in the case of damage-induced filaments: the distribution of added lengths was much broader than those seen for wild type; for example, 15% of divisions occurred without the addition of length between division events. When we assessed SD length as a function of birth length of the filament, we found that there was some correlation between the two. Indeed, the extent of correlation was lesser when compared with steady-state growth, where the length of the daughter is strongly correlated with the birth length of the cell (Figure 2, D and F). To understand whether length added could explain the SD size distribution we observed in the case of asymmetric division in filaments, we analyzed the distributions of length added and SD length at each cell division (Figure 2E). We found that these distributions were significantly different, with length added having a broader distribution when compared with SD sizes. Thus, based on previous studies (Wehrens et al., 2018) and our current observations, this suggested to us that SD size may be more strongly determined by spatial regulation of division events in the filament.
Role of Min-system in division positioning
To characterize the factors contributing to SD production, we first assessed the localization of FtsZ in damage-induced filaments. We observed that the position of FtsZ shifted away from midcell as cell length increased and division occurred one-site-at-a-time in filaments during recovery (Figure 3A and Supplemental Figure S3A). In addition, the number of FtsZ rings did not scale significantly with increasing cell length, with most cells carrying only a single FtsZ-ring and very rarely two (Supplemental Figure S3A, bottom panel). Divisions were completed only one-site-at-a-time even in cephalexin-induced filaments, where 30% cells had more than one visible constriction site (Supplemental Figures S2D and S3D). Consistent with Wehrens et al. (2018), we also observed multiple Min localizations in filaments. Cells had one to two localizations at lengths between 5 and10 μm and this number increased with increasing cell length (Supplemental Figure S3B).
Furthermore, transcript levels of the divisome components did not appear to be perturbed in damage-induced filaments (Supplemental Figure S3C). We hence asked whether factors involved in division regulation contributed in wild-type conditions to SD production during filamentous divisions. For this we assessed the effects of the loss of 1) nucleoid occlusion factor, SlmA, 2) negative regulator of FtsZ polymerization, SulA, and 3) the MinCDE system (de Boer et al., 1989; Mukherjee et al., 1998; Bernhardt and de Boer, 2005; Tonthat et al., 2011; Kreuzer, 2013; Wehrens et al., 2018). We found that deletion of slmA or sulA did not result in an increase in the number of constrictions in filaments (Supplemental Figure S3D). Furthermore, SD size was comparable to that seen in wild-type filaments (Figure 3, B and C), suggesting that these factors may not contribute to division regulation in recovering filaments. In contrast, deletion of the min operon resulted in a more heterogeneous distribution of division site location when compared with wild type. Subsequently, distribution of SD sizes was also found to be more heterogeneous with an increase in frequency of both minicells as well as divisions that could occur near midcell (Figure 3D and Supplemental Figure S3, E and F).
Our observations with min-deleted cells are consistent with previous studies implicating a role for Min-dependent division site regulation in filamentous E. coli. In Wehrens et al. (2018), a Min-dependent division site rule was established for filaments. According to this, there are multiple possible locations of division determined by the multiple nodes of the Min oscillation that vary based on length of the filament, with an equal possibility of division at each position. Given that we observed an effect of min deletion on SD size determination, we asked whether this Min-rule could be recapitulated in the case of damage-induced filaments as well. We plotted the relative position of division for all three durations of DNA damage, binned in three size windows (12–18, 18–26, and 26–33 μm; based on analysis from Wehrens et al., 2018; Figure 3, E–J). Consistent with the Min-rule, we found that filaments of length 12–18 μm showed high frequency of division near the ¼ position. However, in longer filaments (18–33 μm), divisions tended to occur at a higher frequency at the most polar site determined by this Min-rule, with low frequency of midcell division even at filament lengths where such division should be permitted (such as in the size range of 18–26 μm). Interestingly, at filament lengths between 33 and 40 μm, division position seemed more noisy. While division still tended to occur at a high frequency at the most polar division site (0.1–0.15), other sites also had a significant number of division events (e.g., 0.25–0.35). Indeed, this second site is close to the predicted location for possible divisions according to Min-driven division positioning. At these lengths midcell division also occurred; for example, in 15% of filaments in the case of 90 min of damage treatment. Division patterns appear less clearly at longer lengths; however, the number of such long filaments is low across durations of damage. Such long filaments may have accumulated excessive DNA damage due to which recovery dynamics may also be compromised. Indeed, in general, division position seems less precise even at the polar site in damage-induced filaments when compared with division-inhibited filaments (Wehrens et al., 2018); it is possible that this is due to variability in additional factors, such as differences in growth rates. Our observations are largely consistent with previous studies, and suggest that Min is the primary driver of division location in damage-induced filaments as well. However, a significant fraction of divisions occur at the most polar sites and not at other sites predicted by the Min-rule.
Impact of chromosome and terminus segregation on division regulation
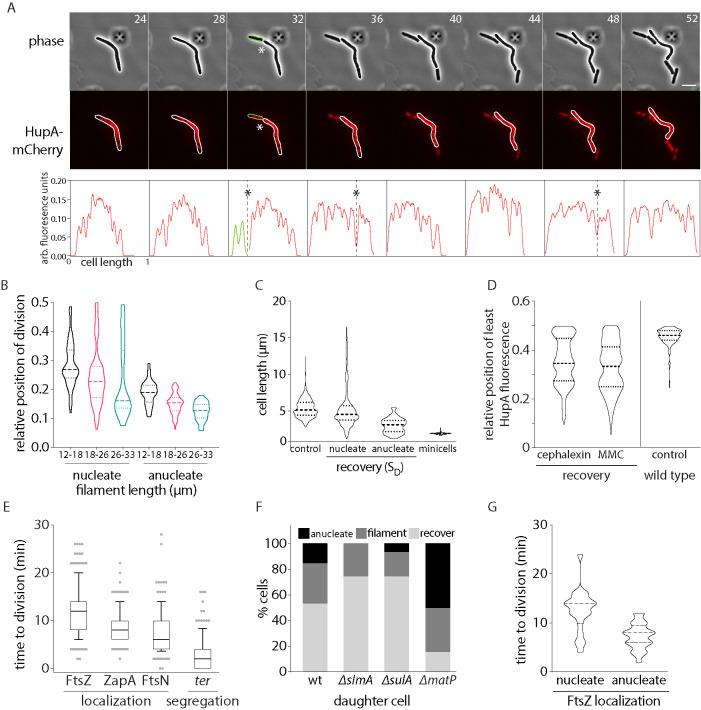
We wondered which additional factors could contribute to higher frequency of polar divisions in the case of damage-induced filaments. Given that chromosomes in DNA damage-induced filaments are no longer segregated (Odsbu and Skarstad, 2014; Vickridge et al., 2017) but chromosome replication continues to proceed (Rudolph et al., 2007; Lloyd and Rudolph, 2016), we asked whether chromosome segregation dynamics in filaments could additionally contribute to control of division, thus influencing the division pattern observed. Hence, we followed chromosomes by imaging the nucleoid-associated protein, HupA (Marceau et al., 2011; Youngren et al., 2014), tagged with mCherry after mitomycin-C (MMC) or cephalexin treatment (Figure 4A).
FIGURE 4:
Impact of chromosome and terminus segregation on division regulation. (A) Representative time-lapse montage of division in cells during recovery. Gray: phase; red, HupA-mCherry (chromosome); scale bar = 5 µm; time in minutes. Fluorescence intensity traces for the cell in montage is provided below. Division sites are marked with “*”. (B) Distribution of relative position of division for filaments between 12 and 18, 18 and 26, and 26 and 33 µm is plotted for nucleate and anucleate (from wild-type and matP backgrounds) divisions (n = 398 [nucleate] and 160 [anucleate]). (C) Cell length distribution for wild-type cells (no perturbation) is plotted. Along with this, cell length distribution of SD during DNA damage recovery is plotted for nucleate cells and anucleate cells. To highlight the distinction between these cell division events and minicell formation, cell length distribution of minicells (from min-deleted cells) is also shown. (n = 1110 [control], 531 [nucleate SD], 41 [anucleate SD], 117 [minicells]). (D) Position of least intensity of HupA fluorescence (gaps between chromosomes) plotted as a function of cell length (from one pole to midcell) in recovering MMC or cephalexin-treated filaments. As reference, these data are also shown for wild-type cells with no damage treatment (control; n = 191 [cephalexin], 476 [MMC], 150 [control]). (E) Distribution of time to division after FtsZ, ZapA, and FtsN localization to division site is plotted. Along with this, time to division after segregation of terminus (ter) during recovery after MMC treatment is also shown (n = 103 [FtsZ], 102 [ZapA], 127 [FtsN], 123 [ter]). (F) Percentage of SD that are anucleate, recover, and filament is plotted for wild type and deletions of slmA, sulA, and matP, during DNA damage recovery (n = 103 [wild type], 105 [slmA], 105 [sulA], 103 [matP]). (G) Time from FtsZ localization to division completion is plotted for divisions that result in nucleated or anucleated SD cells (n = 231 [nucleate], 71 [anucleate]).
When we tracked division in conjunction with chromosome dynamics, we observed that early divisions in filaments resulted in 15 ± 6% of daughter cells that were anucleated (Supplemental Figure S4A). These cells were smaller on average when compared with nucleated cells, but were distinct from minicells (Figure 4C). Anucleate divisions (38%) occurred at the first division event during recovery and this percentage reduced to <7% by the third division. The production of anucleate cells early during division is consistent with Min being the primary driver of divisions, independent of chromosome segregation. When we plotted the relative position of division for anucleate cells, here too division tended to occur at the most polar site as predicted by the Min-rule (consistent with our observations above; Figure 4B). However, division position was less noisy in the case of anucleate division (and more precisely aligned with the Min-rule for the most polar division site) when compared with nucleated divisions. Indeed, anucleate daughter size distribution was also well described by the Min-driven rule for end-point division, with some noise (mean anucleate cell size was 3.4 ± 0.87 µm when compared with a mean cell size of 3.8 ± 0.59 µm as predicted by the Min-rule). However, the Min-rule alone seemed to be insufficient to describe the nucleated division events that had a broader division position and daughter size distribution (mean nucleate cell size was 5.4 ± 2.78 µm vs. 3.8 ± 0.59 µm as predicted by the Min-rule). While we are not sure why division occurs at a higher frequency at polar sites, we propose that this may be driven by the presence of chromosome-free regions closer to the cells poles (Figure 4D and Supplemental Figure S4A), and based on the following additional observations regarding the dynamics of chromosome (and specifically terminus) segregation:
Location of the FtsZ-ring correlated with where the chromosomes were most segregated at the time of division in damage-recovery conditions. Furthermore, chromosomes tended to be better segregated away from midcell (via estimation of location of the least intensity of HupA fluorescence, as a proxy for nucleoid-free regions; Figure 4D and Supplemental Figure S4, B and C).
The terminus region of the chromosome (tracked using the parS-ParB locus labeling system (Nielsen et al., 2006; Espéli et al., 2012) segregated just before or concomitant with division. While the bulk of the chromosome segregated well before division, and divisome components downstream from FtsZ (ZapA and FtsN) localized 8 min and 6 min before division, respectively, a single terminus focus could be observed to persist in the chromosome-free region, where constriction had begun to occur. Constriction was completed and division occurred just before or concomitant with the termini splitting into two foci on either side of the division plane, within 2 min on average (Figure 4E and Supplemental Figure S4, D and E).
Perturbing terminus localization and segregation negatively impacted recovery from DNA damage. Deletion of matP, a key player in modulating terminus dynamics specifically in nondamage conditions (Mercier et al., 2008; Espéli et al., 2012; Nolivos et al., 2016; Lioy et al., 2018), resulted in a significant increase in anucleate cell production, with 50 ± 3% of divisions being anucleated. This is in contrast to wild-type filaments or deletions of slmA or sulA that do not affect terminus segregation, but instead regulate divisome assembly (Figure 4F). Indeed, matP-deleted cells also showed a delay in recovery from DNA damage (as measured by colony-forming units) when compared with wild-type cells (Supplemental Figure S4F).
Based on these observations, we hypothesized that if chromosome segregation has no effect on division dynamics, then the time from divisome assembly to cell division should be the same between anucleated and nucleated divisions. However, if segregation had an effect, then the dynamics would vary between the two types of divisions. In the case of divisions that resulted in the production of anucleated cells, we found that the average period between FtsZ assembly at the division site to cell division was 7 min. In contrast, in the case of nucleated divisions, we noticed that FtsZ localized in the nucleoid-free regions for significantly longer, with an average of 12 min before division (Figure 4G). While the mechanistic details of this process warrants future investigation (see Discussion), these data are in support of a scenario where, in addition to Min-driven division site determination, chromosome (and specifically terminus) segregation contributes to the regulation of cell division in damage-induced filaments.
DISCUSSION
In laboratory conditions, wild-type E. coli maintains a distinct periodicity of cell growth and division that appears to be coupled with chromosome replication and segregation (Donachie, 1968; Hill et al., 2012; Arjes et al., 2014; Campos et al., 2014; Kleckner et al., 2014; Taheri-Araghi et al., 2015; Wallden et al., 2016; Micali et al., 2018). However, it is becoming increasingly evident that bacteria can exist in diverse morphological states, in part dictated by their environmental conditions (Justice et al., 2008; Jonas, 2014; Kysela et al., 2016; Yang et al., 2016; Caccamo and Brun, 2018; MacCready and Vecchiarelli, 2018; Muraleedharan et al., 2018; Heinrich et al., 2019). Even E. coli can become highly filamentous under conditions of stress such as during infection (Justice et al., 2004; Horvath et al., 2011). Transitions into filamentous morphologies are thought to confer several advantages such as avoiding phagocytosis via the host immune response or providing a means to dilute the effects of any inhibitors present in the surroundings (Justice et al., 2004, 2008; Horvath et al., 2011; Yang et al., 2016). Recent reports have also suggested that filamentation (at least in the case of E. coli treated with ciprofloxacin) may be the first step toward the emergence of antibiotic resistance as daughter cells carry mutations making them ciprofloxacin resistant (Bos et al., 2015). Thus, it becomes important to understand how bacterial cells enter and exit these filamentous states to ensure survival under stress.
Here, we find that damage-induced filaments tend to divide asymmetrically, with a high frequency of such divisions resulting in the production of short daughter cells of wild-type size. While the filament itself may or may not recover to wild-type size and still retains damaged DNA, most short daughters generated from a single filament can go on to replicate and divide as wild-type cells. The concept of asymmetric partitioning of cellular components during stress seems to have been coopted by several bacterial systems (Schramm et al., 2019) including Mycobacterium tuberculosis, where one daughter cell inherits the growing pole, while the other has to assemble a growth pole de novo. This results in difference in susceptibility to antibiotic treatment between the two cell types; the daughter cell with the growing pole is more sensitive to cell wall synthesis inhibitors when compared with cells that inherited the nongrowing pole (Aakre and Laub, 2012; Aldridge et al., 2012). The swarmer cells of Proteus mirabilis and Vibrio parahaemolyticus (MacCready and Vecchiarelli, 2018; Muraleedharan et al., 2018) use the Min-system to regulate asymmetric cell division to allow for such division while still preserving the population of filamentous cells. Dim-light stress induces filamentation in the photosynthetic cyanobacterium Synechococcus elongatus, which then divides asymmetrically via positioning by the Min-system (Liao and Rust, 2018). Even in E. coli filaments generated in non-DNA damage conditions, studies have reported poleward division events (Adler and Hardigree, 1965; Begg and Doanachie, 1977; Taschner et al., 1988; Mileykovskaya et al., 1998; Wehrens et al., 2018). It is possible that asymmetric division in damage-induced filaments confers such advantages to short daughters (inheritance of damage-free chromosomes or possible ability of short daughters to swarm away from the stress condition). Taken together, this suggests that switching from midcell to filamentation-based division may be a universal method for cells under stress to ensure viable cell divisions.
How is SD size determined in damage-induced filament divisions? Previous studies have suggested a central role for the Min-system in determining division site rules in filamentous E. coli (Begg and Doanachie, 1977; Taschner et al., 1988; Wehrens et al., 2018). Our observations in min-deleted cells are consistent with the same. However, we observe that midcell divisions may be suppressed at certain filament lengths where such division should be permitted as predicted by the Min-rule seen in division-inhibited filaments (Wehrens et al., 2018). Indeed, division position is more noisy (less accurate) in the case of damage-induced filaments; it is possible that this is due to variability in additional factors in the case of damage-induced filaments, such as differences in growth rates or chromosome segregation dynamics.
We propose the following scenario with regards to polar divisions in damage-induced filaments, based on our results and in line with previous studies: The MinCDE system is the primary contributor to determining the location of divisome assembly. However, time taken to complete division is additionally regulated by chromosome segregation. For example, the disorganized nature of damaged chromosomes, thought to facilitate recombination-based repair (Odsbu and Skarstad, 2014; Vickridge et al., 2017), can add a layer of regulation with chromosome segregation imposing a delay in division completion. The increased frequency of polar division could be a consequence of better chromosome segregation toward cell poles, where chromosome-free spaces are available as a result of molecular crowding (Wu et al., 2019). In most cases cell growth occurs between divisions—it could be that division delay lasts until the Min “division point” and the completion of chromosome “segregation” become aligned. However, in some cases we also observe consecutive divisions without the addition of length in between.
It is currently unclear as to whether and how these two processes (Min-driven division site placement and completion of chromosome segregation) are mechanistically connected. Alternately, it is also possible that divisome positioning and completion of chromosome segregation are concurrent processes that are not molecularly coupled. Indeed, the production of anucleate cells early during filament division as well as when terminus segregation is perturbed are consistent with the idea that segregation is not necessary for cell division, with the Min-system likely being the dominant driver of division (Wehrens et al., 2018). In the case of nucleated divisions, additional mechanisms may contribute to the regulation of division completion, possibly to ensure the completion of chromosome replication and segregation before division, in order to avoid deleterious effects of DNA guillotining (Hendricks et al., 2000). For example, Rudolph et al. (2007) found that cells treated with UV damage continued to initiate new rounds of replication but did not complete the same, likely due to the presence of lesions blocking replication progression. Thus, they observed fewer terminus copies when compared with the origin. The following observations led us to speculate the possibility of such a division checkpoint being regulated by the terminus region of the chromosome during damage recovery (affecting time taken to complete division): 1) In the absence of chromosome segregation, anucleate cells are produced (as also reported by Mulder and Woldringh, 1989). These divisions are faster than nucleated divisions, suggesting that chromosome segregation can impose a delay on timing of division completion. 2) The terminus region of the chromosome persists in nucleoid gaps and segregates just before or concomitant with division. 3) Perturbing terminus organization and segregation via deletion of matP has an impact on outcomes of nucleated cell division and recovery after damage.
The mechanistic basis of such division regulation warrants future investigations. The system characterized in this study provides the ability to assess these rate-limiting steps of division (such as completion of replication) and further probe the mechanisms of asymmetric chromosome segregation that preferentially results in the production of daughter cells devoid of damage. For example, MatP has two independent functions of ter region organization and ter anchoring with the divisome (Bailey et al., 2014; Männik et al., 2016; Nolivos et al., 2016). It would be important to understand which of these activities is specifically necessary in the recovery process. The idea that the terminus region of the chromosome can significantly influence division outcomes has been investigated by several studies in the context of nondamaged E. coli cells (Männik and Bailey, 2015). For example, earlier studies have suggested that perturbing chromosome dimer resolution (via dif mutants) can result in DNA guillotining and subsequent blockage in cell division due to damaged DNA. Such a checkpoint also results in an increase in the production of anucleate cells from filaments (Hendricks et al., 2000). In steady-state conditions, mechanisms including nucleoid occlusion, Min-system, and terminus linkage help coordinate cell division with chromosome segregation to prevent such guillotining (Männik and Bailey, 2015). The ter linkage (via MatP-divisome interactions) also influences Z-ring positioning by acting as a landmark for divisome assembly, thus ensuring precise positioning of the divisome. Lack of this anchoring perturbs division accuracy in steady-state conditions (Espéli et al., 2012; Bailey et al., 2014). In addition to regulating divisome positioning, Buss et al. found that the MatP-FtsZ linkage also slowed down constriction rates, likely contributing to coordination of chromosome segregation with cell division in steady-state conditions (Buss et al., 2015). Thus, our observations with regard to terminus dynamics and MatP-associated phenotypes lend support to the likely importance of this linkage in division control and a possible role for concurrency (Micali et al., 2018) between two mechanisms of division regulation for division accuracy in damage-induced filaments as well. The conservation of asymmetric division in a range of stress-induced bacterial filaments underscores the contribution of this process in facilitating robust exit from the DNA damage checkpoint via the generation of fit, damage-free daughter cells.
MATERIALS AND METHODS
Bacterial strains and growth conditions
Strains and plasmids used in the study are listed in Supplemental Table S1. Oligos used to verify deletions via PCR are listed in Supplemental Table S2. Transductions were conducted with P1 phage following the protocol in Thomason et al. (2007). Transformations were performed following standard chemical competence protocols for E. coli. Cells were grown at 37°C in either rich media (LB: for 1 L dissolved 10 g tryptone, 5 g yeast extract, and 10 g NaCl in double-distilled water) or minimal media (M9-Cas: for 1 L dissolved 5 g glucose, 1 g casamino acids, 1 ml of 0.5% thiamine, 1 ml of 1M MgSO4, and 200 ml 5× M9 salts in double-distilled water). DNA damage was induced with 1 μg/ml MMC for 30, 60, or 90 min (LB) or 90 min (M9-Cas; unless otherwise indicated). Cell division was inhibited using 5 μg/ml cephalexin for 60 min (LB) or 90 min (M9-Cas).
Fluorescence microscopy
Imaging was performed on a wide-field, epifluorescence microscope (Nikon Eclipse Ti-2E, with motorized xy-stage, Z-drift correction), 63X plan apochromat objective (NA 1.41), pE4000 light source (CoolLED), OkoLab incubation chamber, and Hamamatsu Orca Flash 4.0 camera. Images were acquired using the NIS-elements software (version 5.1). Microfluidics imaging was performed using the CellASIC-ONIX2 microfluidic system, temperature controlled CellASIC-ONIX2 Manifold XT, and CellASIC ONIX plate for bacteria cells (B4A; Merck). Details of the imaging and acquisition setting are described here (Raghunathan and Badrinarayanan, 2019).
Time-course and time-lapse imaging
Details of independent repeats of experiments: for damage-recovery (60 min) experiments (with hupA-mCherry), seven biological replicates were performed. ΔslmA experiments were performed twice (biological replicates). FtsZ-GFP/ZapA-GFP recovery time-lapse imaging was conducted independently six times. Wild-type (no damage) time-lapse experiments were performed four times independently. Three independent replicates were performed for all other experiments reported in this study.
For the recovery time course, cells were grown overnight in LB or M9-Cas, back diluted to OD600 = ∼0.01 and allowed to grow to OD600 = ∼0.1. Culture was then treated with MMC or cephalexin for 60 min (LB) or 90 min (M9-Cas). Cells were pelleted, washed with fresh media, resuspended to OD600 = ∼0.1, and then allowed to recover from damage treatment. Cultures were maintained in log phase (OD600 = ∼0.1–0.4) throughout the experiment. For time course, samples for microscopy were collected at the time points indicated. Culture (1 ml) was pelleted, resuspended in 100 μl, and then spotted on a 1% agarose pad and imaged (Chimthanawala and Badrinarayanan, 2019).
For time-lapse experiments, damaged-induced cells were pelleted and washed with fresh media and were either loaded into the microfluidics device or spotted on a 1.5% agarose pad (made with appropriate growth media) and imaged. All time-lapse images were taken every 30 s or 2 min for ∼3 h. For the FtsN imaging experiments, cells were induced with 0.1% rhamnose for 1 h before imaging. For MinD-GFP imaging, cells were induced with 0.2 mM IPTG for 90 min before imaging. Inducers were maintained in the agarose pads or microfluidic plates. Experiments were performed in LB unless otherwise indicated.
RNA sequencing
Protocol for RNA extraction described in Badrinarayanan et al. (2017) was followed. Briefly, cell pellets were collected during the recovery time course at the indicated times. RNA was extracted using the Direct-zol RNA MiniPrep (Zymo, Cat. no. R2052A81) and RNA Clean & Concentrator-25 (Zymo, Cat. no. R1018A82). RNA libraries were prepared using the TruSeq Stranded mRNA Library Preparation kit at NCBS next generation sequencing facility. Libraries were sequenced using the Illumina MiSeq sequencing platform. Raw reads (single end; read length = 50 base pairs) were obtained as .fastq files. The reference genome sequence (.fna) and annotation (.gff) files for the same strain (accession number: NC_000913.3) were downloaded from the National Centre for Biotechnology Information ftp website (“ftp.ncbi.nlm.nih.gov”). The raw read quality was checked using the FastQC software (version v0.11.5). BWA (version 0.7.12-r1039; Li and Durbin, 2009; Burroughs and Aravind, 2016) was used to index the reference genome. Reads with raw read quality ≥20 were aligned using the BWA aln -q option. SAMTOOLS (version 0.1.19-96b5f2294a; Li et al., 2009) was used to filter out the multiply mapped reads. BEDTOOLS (version 2.25.0; Quinlan and Hall, 2010) was used to calculate the reads count per gene using the annotation file (.bed) in a strand-specific manner. Normalized counts were obtained using trimmed mean of M values normalization of EdgeR package. The counts across time were calculated relative to the no damage control. The log2-fold change in expression was plotted from this. Raw data can be accessed via EBI ArrayExpress (EMTAB9696).
Image analysis
Images acquired were visualized and processed using ImageJ (Schindelin et al., 2012, 2015). Data were pooled from two to six replicates per experiment and segmentation was performed using Oufti (Paintdakhi et al., 2016). Foci tracking was accomplished using the Spotfinder Z function of MicrobeTracker (Sliusarenko et al., 2011). For recovery time course, custom MATLAB scripts were used to extract cell length and total fluorescence intensity information for each cell. Anucleate cells were identified using a threshold of 0.02 a.u. for total intensity of HupA-GFP in the cells. For time-lapse analysis, at each division, the longer cell was termed LD and the smaller cell given the identity of SD. A custom MATLAB script was used to the extract the lengths of the LD and SD at each division, the length added between divisions and time between divisions. Daughter cells were classified as “recovers” if at their first division, their length was <10 μm and they divided symmetrically. Cells that were longer at their first division and divided asymmetrically were classified as “filaments.” RecA foci numbers were obtained using Spotfinder Z and combined with cell length information from Oufti. The number of FtsZ rings in a cell were obtained using Spotfinder Z and combined with cell length information from Oufti. For analysis of constrictions and divisions, we used Oufti, which allows subpixel segmentation of phase contrast images of cells in a time-lapse. The method is illustrated in Figure 2 and Supplemental Figure S2. In Figure 2G, a phase profile for a wild-type cell before and after division is plotted. While the profile is, on average, a flat line for most of the time imaged, one can identify a constriction before division (marked with *). Similarly in Figure 2H, the phase profile for a filamentous cell (DNA damage-induced) undergoing division is plotted. If the phase profile deviates by 20–25% of the cell width, it is called a constriction. If the phase profile is discontinuous (with a gap), then it is marked as a division by the segmentation algorithm automatically. For nucleoid tracking, fluorescence intensity profiles of HupA-mCherry, obtained from Oufti, was used. Data was first smoothened using a Savitzgy-Golay filter (Luo et al., 2005). Following this the fluorescence profile was inverted so that the regions of lowest fluorescence now had the highest values. Peaks were determined using the peak prominence function after setting a threshold of half the maximum intensity for each frame. Peaks correspond to lowest fluorescence intensity in the cell. Regions right next to the cell poles were excluded. This information was combined with FtsZ foci tracked using Spotfinder Z to obtain relative positions of the lowest fluorescence intensity in the cell and FtsZ-ring. The formation of FtsZ, ZapA, and FtsN foci, and segregation of the single terminus focus (ParB-GFP) to two foci at the site of division, were scored manually along with time to division.
Supplementary Material
Acknowledgments
The authors are grateful to Thomas Bernhardt, Steven Sandler, Suckjoon Jun, Bill Soederstroem, Fred Boccard, Olivier Espeli, Christian Lesterlin, Ramanujam Srinivasan, and Manjula Reddy for generous sharing of strains and plasmids. The authors acknowledge assistance from Ismath Sadhir in RNA extraction; Nitish Malhotra in RNA-seq analysis; Aditya Jalin, Alex Sam Thomas, and Aalok Varma in writing MATLAB scripts; as well as the Central Imaging and Flow Facility and Next-Generation Genomics Facility. The authors thank anonymous referees, Fagwei Si, Piet de Boer, Raj Ladher, Michael Laub, Tung Le, and members of the A.B. lab for helpful discussions and feedback on the manuscript. This work is supported by funding from National Centre For Biological Sciences–Tata Institute of Fundamental Research (Department of Atomic Energy, Government of India, under project no. 12-R&D-TFR-5.04-0800; A.B. and S.K.), by an Human Frontier Space Program Career Development award (A.B.), and funding from the Simons Foundation (S.K.).
Abbreviations used:
- IPTG
Isopropyl β- d-1-thiogalactopyranoside
- LB
Luria Broth
- LD
long daughter
- M9-Cas
M9–casamino acids
- MMC
mitomycin–C
- NA
numerical aperture
- OD600
optical density at 600 nm wavelength of light
- SD
short daughter.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-08-0547) on October 28, 2020.
REFERENCES
- Aakre CD, Laub MT (2012). Asymmetric cell division: a persistent issue? Dev Cell. 22, 235–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler HI, Hardigree AA (1965). Growth and division of filamentous forms of Escherichia coli. J Bacteriol 90, 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge BB, Fernandez-Suarez M, Heller D, Ambravaneswaran V, Irimia D, Toner M, Fortune SM (2012). Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science 335, 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjes HA, Kriel A, Sorto NA, Shaw JT, Wang JD, Levin PA (2014). Failsafe mechanisms couple division and DNA replication in bacteria. Curr Biol 24, 2149–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badrinarayanan A, Le TBK, Spille J-H, Cisse II, Laub MT (2017). Global analysis of double-strand break processing reveals in vivo properties of the helicase-nuclease complex AddAB. PLoS Genet 13, e1006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MW, Bisicchia P, Warren BT, Sherratt DJ, Männik J (2014). Evidence for divisome localization mechanisms independent of the Min system and SlmA in Escherichia coli. PLOS Genet 10, e1004504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg KJ, Doanachie WD (1977). Growth of the Escherichia coli cell surface. J Bacteriol 129, 1524–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PAJ (2005). SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol Cell 18, 555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J, Zhang Q, Vyawahare S, Rogers E, Rosenberg SM, Austin RH (2015). Emergence of antibiotic resistance from multinucleated bacterial filaments. Proc Natl Acad Sci USA 112, 178–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Aravind L (2016). RNA damage in biological conflicts and the diversity of responding RNA repair systems. Nucleic Acids Res 44, 8525–8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss J, Coltharp C, Shtengel G, Yang X, Hess H, Xiao J (2015). A multi-layered protein network stabilizes the Escherichia coli FtsZ-ring and modulates constriction dynamics. PLOS Genet 11, e1005128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccamo PD, Brun YV (2018). The molecular basis of noncanonical bacterial morphology. Trends Microbiol 26, 191–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos M, Surovtsev IV, Kato S, Paintdakhi A, Beltran B, Ebmeier SE, Jacobs-Wagner C (2014). A constant size extension drives bacterial cell size homeostasis. Cell 159, 1433–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler-Brown D, Schmoller KM, Winetraub Y, Skotheim JM (2017). The adder phenomenon emerges from independent control of pre- and post-start phases of the budding yeast cell cycle. Curr Biol 27, 2774–2783.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimthanawala A, Badrinarayanan A (2019). Live-cell fluorescence imaging of RecN in Caulobactercrescentus under DNA damage. Methods Mol Biol 2004, 239–250. [DOI] [PubMed] [Google Scholar]
- Chung HS, Yao Z, Goehring NW, Kishony R, Beckwith J, Kahne D (2009). Rapid β-lactam-induced lysis requires successful assembly of the cell division machinery. Proc Natl Acad Sci 106, 21872–21877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapa T, Fleurier S, Bredeche M-F, Matic I (2017). The SOS and RpoS regulons contribute to bacterial cell robustness to genotoxic stress by synergistically regulating DNA polymerase Pol II. Genetics 206, 1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer PAJ, Crossley RE, Rothfield LI (1989). A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56, 641–649. [DOI] [PubMed] [Google Scholar]
- Dewachter L, Verstraeten N, Fauvart M, Michiels J (2018). An integrative view of cell cycle control in Escherichia coli. FEMS Microbiol Rev 42, 116–136. [DOI] [PubMed] [Google Scholar]
- Donachie WD (1968). Relationship between cell size and time of initiation of DNA replication. Nature 219, 1077–1079. [DOI] [PubMed] [Google Scholar]
- Espéli O, Borne R, Dupaigne P, Thiel A, Gigant E, Mercier R, Boccard F (2012). A MatP–divisome interaction coordinates chromosome segregation with cell division in E. coli. EMBO J 31, 3198–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli E, Gerdes K (2012). FtsZ-ZapA-ZapB interactome of Escherichia coli. J. Bacteriol 194, 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris LK, Theriot JA (2016). Relative rates of surface and volume synthesis set bacterial cell size. Cell 165, 1479–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris LK, Theriot JA (2018). Surface area to volume ratio: a natural variable for bacterial morphogenesis. Trends Microbiol 26, 815–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich K, Leslie DJ, Morlock M, Bertilsson S, Jonas K (2019). Molecular basis and ecological relevance of caulobacter cell filamentation in freshwater habitats. MBio 10, 10.1128/mBio.01557-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks EC, Szerlong H, Hill T, Kuempel P (2000). Cell division, guillotining of dimer chromosomes and SOS induction in resolution mutants (dif, xerC and xerD) of Escherichia coli. Mol Microbiol 36, 973–981. [DOI] [PubMed] [Google Scholar]
- Hill NS, Kadoya R, Chattoraj DK, Levin PA (2012). Cell size and the initiation of DNA replication in bacteria. PLoS Genet 8, e1002549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath DJ, Li B, Casper T, Partida-Sanchez S, Hunstad DA, Hultgren SJ, Justice SS (2011). Morphological plasticity promotes resistance to phagocyte killing of uropathogenic Escherichia coli. Microbes Infect 13, 426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas K (2014). To divide or not to divide: control of the bacterial cell cycle by environmental cues. Curr Opin Microbiol 18, 54–60. [DOI] [PubMed] [Google Scholar]
- Justice SS, Hung C, Theriot JA, Fletcher DA, Anderson GG, Footer MJ, Hultgren SJ (2004). Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc Natl Acad Sci USA 101, 1333–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice SS, Hunstad DA, Cegelski L, Hultgren SJ (2008). Morphological plasticity as a bacterial survival strategy. Nat Rev Microbiol 6, 162–168. [DOI] [PubMed] [Google Scholar]
- Kantor GJ, Deering RA (1966). Ultraviolet radiation studies of filamentous Escherichia coli B. J Bacteriol 92, 1062–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner NE, Chatzi K, White MA, Fisher JK, Stouf M (2018). Coordination of growth, chromosome replication/segregation, and cell division in E. coli. Front Microbiol 9, 1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N, Fisher JK, Stouf M, White MA, Bates D, Witz G (2014). The bacterial nucleoid: nature, dynamics and sister segregation. Curr Opin Microbiol 22, 127–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer KN (2013). DNA damage responses in prokaryotes: regulating gene expression, modulating growth patterns, and manipulating replication forks. Cold Spring Harb Perspect Biol 5, a012674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kysela DT, Randich AM, Caccamo PD, Brun YV (2016). Diversity takes shape: understanding the mechanistic and adaptive basis of bacterial morphology. PLoS Biol 14, e1002565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert A, Vanhecke A, Archetti A, Holden S, Schaber F, Pincus Z, Laub MT, Goley E, Manley S (2018). Constriction rate modulation can drive cell size control and homeostasis in C. crescentus. iScience. 4, 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesterlin C, Ball G, Schermelleh L, Sherratt DJ (2014). RecA bundles mediate homology pairing between distant sisters during DNA break repair. Nature 506, 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Rust MJ (2018). The Min oscillator defines sites of asymmetric cell division in cyanobacteria during stress recovery. Cell Syst. 7, 471–481.e6. [DOI] [PubMed] [Google Scholar]
- Lioy VS, Cournac A, Marbouty M, Duigou S, Mozziconacci J, Espéli O, Boccard F, Koszul R (2018). Multiscale structuring of the E. coli chromosome by nucleoid-associated and condensin proteins. Cell 172, 771–783.e18. [DOI] [PubMed] [Google Scholar]
- Lloyd RG, Rudolph CJ (2016). 25 years on and no end in sight: a perspective on the role of RecG protein. Curr Genet 62, 827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Ying K, Bai J (2005). Savitzky-Golay smoothing and differentiation filter for even number data. Signal Process 85, 1429–1434. [Google Scholar]
- MacCready JS, Vecchiarelli AG (2018). In long bacterial cells, the Min system can act off-center. Mol. Microbiol. 109, 268–272. [DOI] [PubMed] [Google Scholar]
- Männik J, Bailey MW (2015). Spatial coordination between chromosomes and cell division proteins in Escherichia coli. Front Microbiol. 6, 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Männik J, Castillo DE, Yang D, Siopsis G, Männik J (2016). The role of MatP, ZapA and ZapB in chromosomal organization and dynamics in Escherichia coli. Nucleic Acids Res 44, 1216–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau AH, Bahng S, Massoni SC, George NP, Sandler SJ, Marians KJ, Keck JL (2011). Structure of the SSB-DNA polymerase III interface and its role in DNA replication. EMBO J 30, 4236–4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Petit M-A, Schbath S, Robin S, El Karoui M, Boccard F, Espéli O (2008). The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell 135, 475–485. [DOI] [PubMed] [Google Scholar]
- Micali G, Grilli J, Osella M, Lagomarsino MC (2018). Concurrent processes set E. coli cell division. Sci Adv 4, eaau3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileykovskaya E, Sun Q, Margolin W, Dowhan W (1998). Localization and function of early cell division proteins in filamentous Escherichia coli cells lacking phosphatidylethanolamine. J. Bacteriol 180, 4252–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo AH, Burkholder WF (2010). YneA, an SOS-induced inhibitor of cell division in Bacillus subtilis, is regulated posttranslationally and requires the transmembrane region for activity. J Bacteriol 192, 3159–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modell JW, Hopkins AC, Laub MT (2011). A DNA damage checkpoint in Caulobacter crescentus inhibits cell division through a direct interaction with FtsW. Genes Dev 25, 1328–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modell JW, Kambara TK, Perchuk BS, Laub MT (2014). A DNA damage-induced, SOS-independent checkpoint regulates cell division in Caulobactercrescentus. PLoS Biol 12, e1001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Cao C, Lutkenhaus J (1998). Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc Natl Acad Sci USA 95, 2885–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder E, Woldringh CL (1989). Actively replicating nucleoids influence positioning of division sites in Escherichia coli filaments forming cells lacking DNA. J Bacteriol 171, 4303–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraleedharan S, Freitas C, Mann P, Glatter T, Ringgaard S (2018). A cell length-dependent transition in MinD-dynamics promotes a switch in division-site placement and preservation of proliferating elongated Vibrio parahaemolyticus swarmer cells. Mol. Microbiol 109, 365–384. [DOI] [PubMed] [Google Scholar]
- Nielsen HJ, Ottesen JR, Youngren B, Austin SJ, Hansen FG (2006). The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves: E. coli chromosome segregation. Mol Microbiol 62, 331–338. [DOI] [PubMed] [Google Scholar]
- Nolivos S, Upton AL, Badrinarayanan A, Müller J, Zawadzka K, Wiktor J, Gill A, Arciszewska L, Nicolas E, Sherratt D (2016). MatP regulates the coordinated action of topoisomerase IV and MukBEF in chromosome segregation. Nat Commun 7, 10466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odsbu I, Skarstad K (2014). DNA compaction in the early part of the SOS response is dependent on RecN and RecA. Microbiology 160, 872–882. [DOI] [PubMed] [Google Scholar]
- Paintdakhi A, Parry B, Campos M, Irnov I, Elf J, Surovtsev I, Jacobs-Wagner C (2016). Oufti: an integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Mol Microbiol 99, 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan S, Badrinarayanan A (2019). Tracking bacterial chromosome dynamics with microfluidics-based live cell imaging. Methods Mol Biol 2004, 223–238. [DOI] [PubMed] [Google Scholar]
- Rajendram M, Zhang L, Reynolds BJ, Auer GK, Tuson HH, Ngo KV, Cox MM, Yethiraj A, Cui Q, Weibel DB (2015). Anionic phospholipids stabilize RecA filament bundles in Escherichia coli. Mol Cell 60, 374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Lamothe R, Sherratt DJ (2019). The bacterial cell cycle, chromosome inheritance and cell growth. Nat Rev Microbiol 17, 467–478. [DOI] [PubMed] [Google Scholar]
- Rolinson GN (1980). Effect of β-Lactam antibiotics on bacterial cell growth rate. Microbiology 120, 317–323. [DOI] [PubMed] [Google Scholar]
- Rudolph CJ, Upton AL, Briggs GS, Lloyd RG (2010). Is RecG a general guardian of the bacterial genome? DNA Repair (Amst) 9, 210–223. [DOI] [PubMed] [Google Scholar]
- Rudolph CJ, Upton AL, Harris L, Lloyd RG (2009). Pathological replication in cells lacking RecG DNA translocase. Mol Microbiol 73, 352–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph CJ, Upton AL, Lloyd RG (2007). Replication fork stalling and cell cycle arrest in UV-irradiated Escherichia coli. Genes Dev 21, 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Rueden CT, Hiner MC, Eliceiri KW (2015). The ImageJ ecosystem: an open platform for biomedical image analysis. Mol Reprod Dev 82, 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm FD, Schroeder K, Alvelid J, Testa I, Jonas K (2019). Growth-driven displacement of protein aggregates along the cell length ensures partitioning to both daughter cells in Caulobacter crescentus. Mol Microbiol 111, 1430–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si F, Le Treut G, Sauls JT, Vadia S, Levin PA, Jun S (2019). Mechanistic origin of cell-size control and homeostasis in bacteria. Curr Biol 29, 1760–1770.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C (2011). High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol Microbiol 80, 612–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soifer I, Robert L, Amir A (2016). Single-cell analysis of growth in budding yeast and bacteria reveals a common size regulation strategy. Curr Biol 26, 356–361. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Pangborn J, Kilgore WW (1967). Filamentous cells of Escherichia coli formed in the presence of mitomycin. J Bacteriol 93, 683–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri-Araghi S, Bradde S, Sauls JT, Hill NS, Levin PA, Paulsson J, Vergassola M, Jun S (2015). Cell-size control and homeostasis in bacteria. Curr Biol 25, 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner PE, Huls PG, Pas E, Woldringh CL (1988). Division behavior and shape changes in isogenic ftsZ, ftsQ, ftsA, pbpB, and ftsE cell division mutants of Escherichia coli during temperature shift experiments. J Bacteriol 170, 1533–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason LC, Costantino N, Court DL (2007). E. coli genome manipulation by P1 transduction. Curr Protoc Mol Biol Chapter 1:Unit 1.17. 10.1002/0471142727.mb0117s79. [DOI] [PubMed] [Google Scholar]
- Tonthat NK, Arold ST, Pickering BF, Van Dyke MW, Liang S, Lu Y, Beuria TK, Margolin W, Schumacher MA (2011). Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. EMBO J 30, 154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang M-J, Bernhardt TG (2015). Guiding divisome assembly and controlling its activity. Curr Opin Microbiol 24, 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickridge E, Planchenault C, Cockram C, Junceda IG, Espéli O (2017). Management of E. coli sister chromatid cohesion in response to genotoxic stress. Nat. Commun 8, 14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallden M, Fange D, Lundius EG, Baltekin Ö, Elf J (2016). The synchronization of replication and division cycles in individual E. coli cells. Cell 166, 729–739. [DOI] [PubMed] [Google Scholar]
- Wehrens M, Ershov D, Rozendaal R, Walker N, Schultz D, Kishony R, Levin PA, Tans SJ (2018). Size laws and division ring dynamics in filamentous Escherichia coli cells. Curr Biol 28, 972–979.e5. [DOI] [PubMed] [Google Scholar]
- Wu F, Swain P, Kuijpers L, Zheng X, Felter K, Guurink M, Solari J, Jun S, Shimizu TS, Chaudhuri D, et al. (2019). Cell boundary confinement sets the size and position of the E. coli chromosome. Curr Biol 29, 2131–2144.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DC, Blair KM, Salama NR (2016). Staying in shape: the impact of cell shape on bacterial survival in diverse environments. Microbiol Mol Biol Rev 80, 187–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngren B, Nielsen HJ, Jun S, Austin S (2014). The multifork Escherichia coli chromosome is a self-duplicating and self-segregating thermodynamic ring polymer. Genes Dev 28, 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.