Abstract
Background
SARS-CoV-2 molecular diagnostics is facing material shortages and long turnaround times due to exponential increase of testing demand.
Objective
We evaluated the analytic performance and handling of four rapid Antigen Point of Care Tests (AgPOCTs) I-IV (Distributors: (I) Roche, (II) Abbott, (III) MEDsan and (IV) Siemens).
Methods
100 RT-PCR negative and 84 RT-PCR positive oropharyngeal swabs were prospectively collected and used to determine performance and accuracy of these AgPOCTs. Handling was evaluated by 10 healthcare workers/users through a questionnaire.
Results
The median duration from symptom onset to sampling was 6 days (IQR 2–12 days). The overall respective sensitivity were 49.4 % (CI95 %: 38.9–59.9), 44.6 % (CI95 %: 34.3–55.3), 45.8 % (CI95 %: 35.5–56.5) and 54.9 % (CI95 %: 43.4−65.9) for tests I, II, III and IV, respectively. In the high viral load subgroup (containing >106 copies of SARS-CoV-2 /swab, n = 26), AgPOCTs reached sensitivities of 92.3 % or more (range 92.3 %–100 %). Specificity was 100 % for tests I, II (CI95 %: 96.3–100 for both tests) and IV (CI95 %: 96.3–100) and 97 % (CI95 %: 91.5–98.9) for test III. Regarding handling, test I obtained the overall highest scores, while test II was considered to have the most convenient components. Of note, users considered all assays, with the exception of test I, to pose a significant risk for contamination by drips or spills.
Discussion
Besides some differences in sensitivity and handling, all four AgPOCTs showed acceptable performance in high viral load samples. However, due to the significantly lower sensitivity compared to RT-qPCR, a careful consideration of pro and cons of AgPOCT has to be taken into account before clinical implementation.
Abbreviations: RT-qPCR, Reverse transcription-polymerase chain reaction, AgPOCT
Keywords: Accuracy, Handling, SARS-CoV-2, Rapid antigen test AgPOCT
1. Introduction
The SARS-CoV-2 pandemic continues to spread at accelerating pace [1], posing an unprecedented challenge for health care systems around the globe. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis remains the gold standard for diagnosis of SARS-CoV-2 infection [2]; however, limited supply of material and specialized personnel, as well as frequent reporting delays bring about the need for alternative testing formats. Rapid Antigen Point of Care Tests (AgPOCTs) represent one such alternative and have been pushed into the market by countless distributors in recent months. The tally of CE-cleared AgPOCT products has mounted to just over 200 during this time [3], leaving potential users mostly in the dark about their performance in real-world situations as independent comparison studies were unable to keep pace with the flood of novel tests [4,5].
2. Objective
The aim of this study was to evaluate and compare handling, analytical- and clinical performance of four commercial rapid antigen point of care tests (AgPOCT) distributed by established diagnostics firms.
3. Methods
3.1. Clinical samples and ethics
Single center non-interventional diagnostic multiple-gate study. Respiratory samples were oropharyngeal or nasopharyngeal swabs collected using Universal Transport Medium (UTM) based collection kits by Copan (Italy, Brescia) or Iclean (Shenzhen, China) and prospectively collected following routine diagnostics at the University Medical Center Hamburg-Eppendorf (during August through November 2020) from patients hospitalized with suspected or known COVID-19. Once the required number of positive RT-qPCR SARS-CoV-2 samples was collected, a further 100 RT-qPCR negative SARS-CoV-2 samples were randomly selected to serve as negative control. This work was conducted in accordance with §12 of the Hamburg hospital law (§12 HmbKHG). The use of anonymized samples was approved by the ethics committee, Freie und Hansestadt Hamburg, PV5626.
AgPOCT and RT-qPCR procedures: Four different CE-labeled AgPOCT kits were compared (Table 1 ). Swabs supplied with the AgPOCT kits were immersed in patient oropharyngeal/nasopharyngeal samples for approximately 10 s before all further steps of the tests were carried out according to instructions by manufacturers. The results of the AgPOCTs were read by two unblinded operators. It has to be noted, that this particular protocol of dipping nasopharyngeal swabs into UTM samples is a deviation from the recommended sample matrix and or handling recommendations by the manufacturer.
Table 1.
Main characteristics of four rapid SARS-CoV-2 rapid antigen tests (according to the manufacturer).
| Test I | Test II | Test III | Test IV | |
|---|---|---|---|---|
| Description | SARS-CoV-2 Rapid Antigen Test (Roche) | COVID-19 Rapid Test Device (Abbott) | MEDsan SARS-CoV-2 Antigen Rapid Test | CLINITEST Rapid COVID.19 Antigen Test |
| Distributor | Roche Diagnostics | Abbott Rapid Diagnostics | MEDsan GmbH | Siemens |
| Manufacturer | SD Biosensor | Panbio Ltd. | MEDsan GmbH | Zhejiang Orient Biotech Co. |
| Country of origin | South Korea | Australia | Germany | China |
| Certification | CE-IVD | CE-IVD | CE-IVD | CE-IVD |
| Specimen | NP | NP | NP | NP |
| Limit of detection (manufacturer) | 3,1 × 102,2 TCID50/mL | 2,5 × 102 TCID50/mL | 1,4 × 102 TCID50/mL | 1,1 × 102 TCID50/mL |
| Volume applied into cassette | 3 drops | 5 drops | 2 drops | 4 drops |
| Incubation | 15−30 minutes | 15−20 minutes | 15−20 minutes | 15−20 minutes |
| Readout | Visual: colored band | Visual: colored band | Visual: colored band | Visual: colored band |
| UTM protocol | Reported | Not specified | Not specified | Not specified |
| Cross-reactivity against other human respiratory virus evaluated and excluded | Reported | Reported | Reported | Reported |
N-protein: Nucleocapsid protein Ag.
NP: nasopharyngeal swab.
TCID50: Median tissue culture infective dose.
Based on previous experience, we postulated that roughly 100 μL of UTM sample are carried over by the procedure described above. To allow for a meaningful clinical correlation, ‘effective’ viral loads/swab were calculated by applying a factor of 0.1 to all PCR measurements from original samples. This was done in an effort to generate results more representative of a real-life testing procedure. A commercial SARS-CoV-2 RT-qPCR assay (Roche cobas SARS-CoV-2 IVD) was used as reference, performed directly from UTM samples.
For limit of detection experiments, a serial dilution of cell-free virus solution containing SARS-CoV-2 strain HH-1 [6] in UTM was prepared and subjected to all four tests with five repeats per dilution step. As RT-qPCR reference method, the cobas6800 SARS-CoV-2 IVD assay was used in conjunction with quantitative external control material by Instand e.V. (Düsseldorf, Germany) to allow for absolute quantification, as previously described [7,8]. To evaluate the ease of handling and implementation of AgPOCTs into clinical routine we conducted a user survey employing a questionnaire (see supplementary Table S1) with 10 participants representing different clinical specialties and professions (3 Intensive Care Unit (ICU) medical doctors, 2 ICU nurses, 3 microbiologists and 2 lab technicians). All users had no prior experience with SARS-CoV-2 AgPOCT, but have handled other lateral flow tests in the past. Every user received the complete set of materials for every test, including the instructions manual. Questionnaires were filled in anonymously and independently by each participant. A 5-point Likert scale (1 = “do not agree at all” to 5 = “absolutely agree”) was used to evaluate for different test dimensions. Statistical analysis was performed with STATA (version 15) and GraphPad Prism (version 86 9.0.0).
4. Results
4.1. Analytical and clinical performance
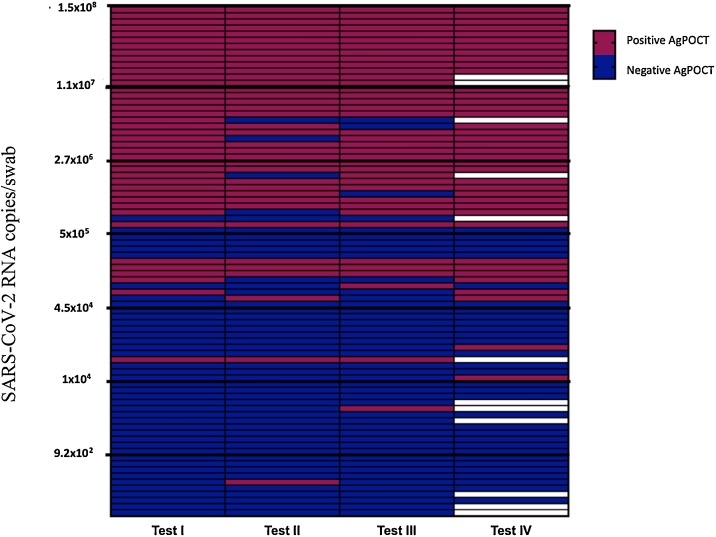
In total 100 RT-qPCR negative and 84 positive respiratory samples were included in this study (test IV was performed on only 72 positive respiratory samples; Fig. 1 ).56.5 % (26/ 46, unknown 38) of specimens were taken within one week of symptom onset. The median duration from symptom onset to sampling was 6 days (IQR 2–12 days). Median viral load/ swab for the AgPOCT was 3.8 × 105 copies/swab (IQR 5.4 × 103 – 3.8 × 106 copies/swab). Clinical sensitivity was 49.4 % (CI95: 38.9 %–59.9 %), 44.6 % (CI95: 34.3 %–55.3 %), 45.8 % (CI95: 35.5 %–56.5 %) and 54.9 % (CI95: 43.4 %–65.9 %) for the tests I, II, III and IV, respectively (Table 2 ). Limit of Detection (LOD) using cell culture derived SARS-CoV-2 virus was in line with these results showing similar analytical performance with best performance for test IV (see suppl. Fig. S1). All assays showed a specificity of 100 %, with the exception of test III (97 % (CI95: 91.5 %–98.9 %), see Table 2). In a set of samples that all contained > 106 copies/swab (n = 32 for test I,II and III and n = 28 for test IV; median duration from symptom onset to sampling 6 days (IQR 2–9)), clinical sensitivity rose to 100 % (CI95: 87 %–100 %), 92.3 % (CI95: 75.8 %–97.8 %), 92.3 % (CI95: 75.8 %–97.8 %) and 100 % (CI95: 85.7 %–100 %) for tests I, II, III and IV, respectively.
Fig. 1.
AgPOCT results in relation to SARS-CoV-2 RNA copies/swab. Purple fields represent positive and blue fields negative AgPOCT results. Blank fields correspond to cases where samples were not tested for the corresponding assay (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
Table 2.
Performance of four rapid antigen tests for SARS-CoV-2 compared to RT-qPCR.
| Test | Sensitivity % (CI95) | Specificity % (CI95) | Accuracy % | Kappa coefficient |
|---|---|---|---|---|
| I (n positive = 84; n negative = 100) | 49.4 (38.9−59.9) | 100 (96.3–100) | 77.1 | 0.52 |
| II (n positive = 84; n negative = 100) | 44.6 (34.3−55.3) | 100 (96.3–100) | 74.8 | 0.46 |
| III (n positive = 84; n negative = 100) | 45.8 (35.5−56.5) | 97 (91.5–98.9) | 73.7 | 0.45 |
| IV (n positive = 72; n negative = 100) | 54.9 (43.4−65.9) | 100 (96.3–100) | 81.2 | 0.58 |
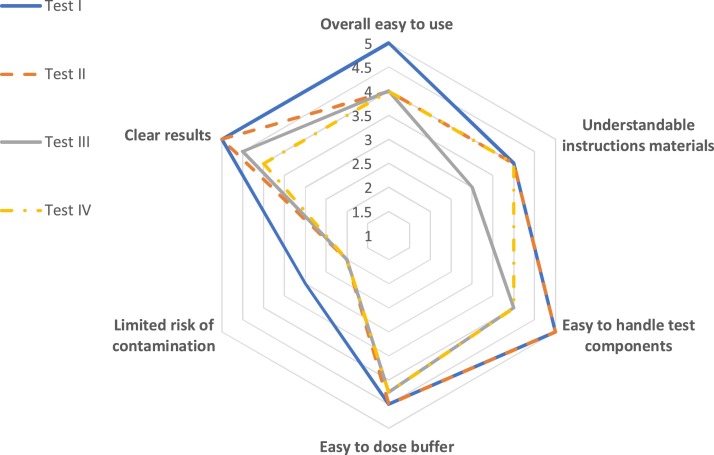
4.2. Handling
The median of the results for each item is illustrated in Fig. 2 . Test I was considered the overall easiest to usefollowed by test II. Test III scored lowest overall. Of note, users considered all assays to pose a significant risk for contamination by drips or spills (see Fig. 2).
Fig. 2.
Evaluation of the handling of four AgPOCTs. Scores vary from 1 (“do not agree at all”) to 5 (“absolutely agree”).
5. Discussion
Here we report a head-to-head comparison of performance and handling for four rapid AgPOCTs of several major distributors in Europe, using the latest version of CE-labeled kits. All tests were able to detect 106 or more copies/swab with high reliability (95 %), implying that patients with high viral loads can be identified with acceptable accuracy. This is in line with previous studies [4,5].
Relative clinical sensitivity of rapid SARS-CoV-2 antigen tests has repeatedly fallen short of optimistic manufacturer claims [4,[9], [10], [11]], in large parts due to differences in patient characteristics in respective cohorts and artificial conditions when performing AgPOCT. This also applies to this study, as e.g. antigen tests were performed from clinical UTM samples. Still, the lack of an amplification step inherently entails significantly higher analytic limits when compared to RT-qPCR, which is expected to be 100–1000 times more sensitive than an AgPOCT. However, in the context of increased frequency of testing, lower sensitivity tests may be just as effective as highly sensitive ones in identifying individual infections and outbreaks, as suggested by mathematical modeling studies [12].
All AgPOCT evaluated in this study showed largely comparable clinical and analytical sensitivity, with the exception of Test IV (Siemens). The slight edge Test IV had in LoD experiments translated into a moderately increased positive agreement with RT-qPCR (of 54 %) compared to the other tests (lower than 50 %). It has to be noted that fewer clinical samples were tested with Test IV as it only became available when experiments were already underway, thus providing only an incomplete dataset to compare with the other tests. Furthermore, the limited sensitivity of the assays can also be justified by the fact that half of the samples were collected at least one week after symptom onset, as shown in other studies [13].
Besides analytical properties, handling and practical application were evaluated as part of this study. While all AgPOCTs were deemed “easy to use”, Test I was considered slightly better than the rest, while Test II was considered to have the most convenient components. We were surprised to notice that most users reported a high risk of contamination (by drips or spills). This aspect should not be underestimated in particular considering AgPOCT are to be used outside the health care sector and might also be performed by untrained personnel lacking experience in dealing with infectious materials. Especially in a context where test performance is largely identical, practical aspects should not be neglected when choosing which test to employ, as minor inconveniences in performing a single test can add up to massive delays when performing them in large numbers.
In conclusion, we present analytical and clinical evaluation of four rapid SARS-CoV-2 antigen tests, as well as a user survey for practical application. All tests demonstrated largely similar performance and are expected to detect high viral loads with adequate reliability. RT-qPCR remains the gold standard to definitively confirm or rule out infections due to its significantly higher sensitivity and specificity.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jcv.2021.104782.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Covid-19: Is a second wave hitting Europe? | The BMJ, (n.d.). https://www.bmj.com/content/371/bmj.m4113 (accessed November 19, 2020). [DOI] [PubMed]
- 2.Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases, (n.d.). https://www.who.int/publications-detail-redirect/10665-331501 (accessed November 19, 2020).
- 3.Liste der Antigentests, (n.d.). https://antigentest.bfarm.de/ords/antigen/r/antigentests-auf-sars-cov-2/liste-der-antigentests?session=21375807250974 (accessed November 24, 2020).
- 4.Weitzel T., Legarraga P., Iruretagoyena M., Pizarro G., Vollrath V., Araos R., Munita J.M., Porte L. Head-to-head comparison of four antigen-based rapid detection tests for the diagnosis of SARS-CoV-2 in respiratory samples. BioRxiv. 2020 doi: 10.1101/2020.05.27.119255. 2020.05.27.119255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comparison of seven commercial SARS-CoV-2 rapid Point-of-Care Antigen tests | medRxiv, (n.d.). https://www.medrxiv.org/content/10.1101/2020.11.12.20230292v1.article-info (accessed November 24, 2020).
- 6.Pfefferle S., Reucher S., Nörz D., Lütgehetmann M. Evaluation of a quantitative RT-PCR assay for the detection of the emerging coronavirus SARS-CoV-2 using a high throughput system. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.9.2000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nörz D., Fischer N., Schultze A., Kluge S., Mayer-Runge U., Aepfelbacher M., Pfefferle S., Lütgehetmann M. Clinical evaluation of a SARS-CoV-2 RT-PCR assay on a fully automated system for rapid on-demand testing in the hospital setting. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2020;128:104390. doi: 10.1016/j.jcv.2020.104390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nörz D., Frontzek A., Eigner U., Oestereich L., Wichmann D., Kluge S., Fischer N., Aepfelbacher M., Pfefferle S., Lütgehetmann M. Pushing beyond specifications: evaluation of linearity and clinical performance of the cobas 6800/8800 SARS-CoV-2 RT-PCR assay for reliable quantification in blood and other materials outside recommendations. J. Clin. Virol. 2020;132:104650. doi: 10.1016/j.jcv.2020.104650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mak G.C., Cheng P.K., Lau S.S., Wong K.K., Lau C., Lam E.T., Chan R.C., Tsang D.N. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020;129:104500. doi: 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scohy A., Anantharajah A., Bodéus M., Kabamba-Mukadi B., Verroken A., Rodriguez-Villalobos H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2020;129:104455. doi: 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinnes J., Deeks J.J., Adriano A., Berhane S., Davenport C., Dittrich S., Emperador D., Takwoingi Y., Cunningham J., Beese S., Dretzke J., Ferrante di Ruffano L., Harris I.M., Price M.J., Taylor-Phillips S., Hooft L., Leeflang M.M., Spijker R., Van den Bruel A. Cochrane COVID-19 Diagnostic Test Accuracy Group, Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2020;8 doi: 10.1002/14651858.CD013705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mina M.J., Parker R., Larremore D.B. Rethinking Covid-19 test sensitivity — a strategy for containment. N. Engl. J. Med. 2020 doi: 10.1056/NEJMp2025631. [DOI] [PubMed] [Google Scholar]
- 13.Bullard J., Dust K., Funk D., Strong J.E., Alexander D., Garnett L., Boodman C., Bello A., Hedley A., Schiffman Z., Doan K., Bastien N., Li Y., Van Caeseele P.G., Poliquin G. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020 doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.