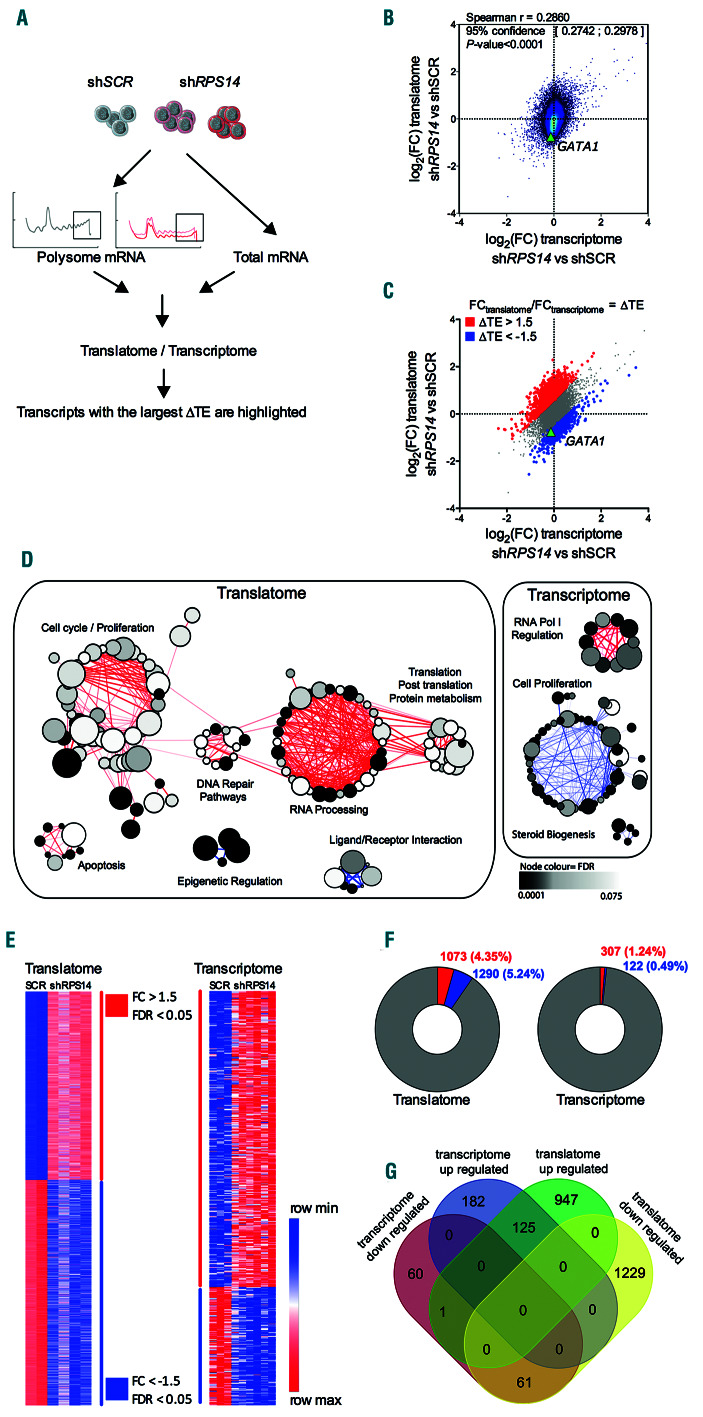
Figure 2.
Selective translation in UT-7/EPO shRPS14 cell lines. (A) Schematic workflow showing RNA extraction from actively translating polysomes (translatome) or from whole cell lysates (transcriptome) in shRPS14 640, shRPS14 641 and shSCR cells to be used for microarray analysis. Comparative analysis of the translatome related to the transcriptome and the determination of transcripts with the largest translational efficiencies variation (ΔTE) are shown. (B) 2D density plot of the log2(foldchange, FC) values obtained for the translatome and transcriptome. A Spearman test (r coefficient with 95% confidence interval and P-value) was used for correlation analysis. (C) Post-transcriptionally regulated components with an (FCtranslatome/FCtranscriptome) ratio >1.5 or <1/1.5 were overlaid as red or blue dots, respectively. (D) Gene set enrichment analysis (GSEA) of differentially expressed genes in the translatome and transcriptome. Enriched cellular programs are indicated using the Enrichment Map App of Cytoscape software (P-value cut-off = 0.005, q-value cut-off = 0.1). Nodes represent the gene sets that were enriched at the top or bottom ranking of the differentially expressed genes. Node size corresponds to the number of genes in the set. Edges indicate overlaps between gene sets and the line thickness indicates the similarity coefficient between gene sets in red for less impacted and blue for downregulated genes. The node color depends on the false discovery rate (FDR) value as shown on the scale. Clusters of nodes were determined with the Autoannotate App and are represented by the circles. (E) Heatmaps representing the significantly up- or downregulated transcripts in the translatome and transcriptome with a P-value cut-off <0.05, a calculated q-value (FDR) in significance analysis microarrays (SAM) ≤0.05 and a minimum fold-change of >1.5 or <1/1.5. The color scale representing the minimum and maximum expression per row is indicated. (F) Numbers of significantly up- or downregulated transcripts shown as doughnuts. (G) Venn diagram showing the overlap between the significantly less impacted or more downregulated transcripts in the translatome and transcriptome.