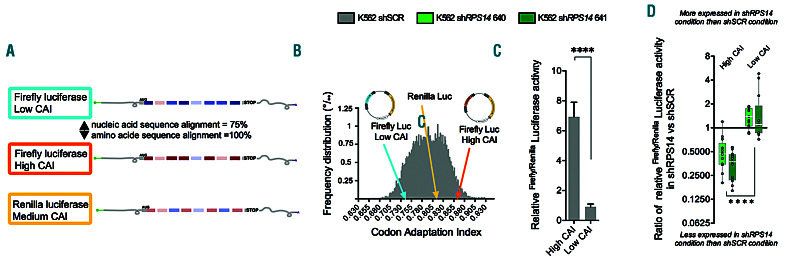
Figure 4.
Validation of codon bias as a determinant of translation selectivity. K562 shRPS14640, shRPS14641 and shSCR cells were used in dual luciferase reporter assays. (A) Schematic representation of the luciferase codon contents. (B) Frequency distribution of the codon adaptation index (CAI) values among all human transcripts. The CAI of firefly and renilla luciferases are indicated. (C) Firefly/renilla luciferase activity in K562 shSCR cells comparing firefly luciferase with a high versus that with a low CAI. (D) Ratios of firefly/renilla luciferase activity in K562 shRPS14 to K562 shSCR comparing firefly luciferase with a high or low CAI (n=13). Medians are shown as white horizontal bars, boxes represent first and third quartiles, and whiskers represent minimal and maximal values. Light green denotes shRPS14640 and dark green indicates shRPS14641. Unpaired two-tailed Student t-test: **P<0.01; ****P<0.0001.