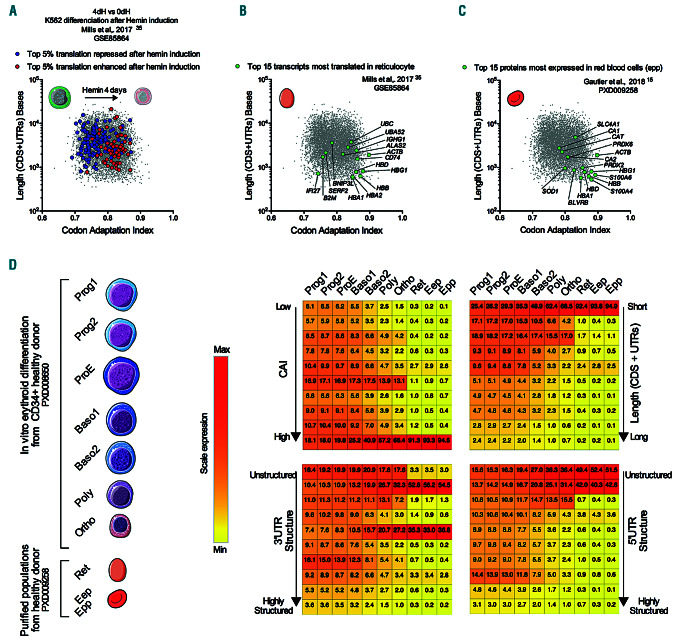
Figure 7.
The codon adaptation index, coding sequence length and thermodynamic characteristics of the untranslated regions (UTR) in an mRNA are key determinants of its translation during normal erythropoiesis. (A-C) Bi-parametric scatter plot in a log scale of the codon adaptation index (CAI) and length (coding sequence [CDS]+UTR) of all human transcripts denoted by gray dots. (A) The top five percentile transcripts which are the more or the less translated after hemin induction of differentiation in K562 model (Mills et al.,35 GSE85864) are overlaid in red or blue, respectively. dH: day post hemin. (B) The top 15 transcripts most translated mRNAs in reticulocyte (Mills et al.,35 GSE85864) overlaid in green (C) The top 15 most expressed proteins in red blood cells (Gautier et al.,15 PXD009258) overlaid in green. (D) Dynamic repartition of proteins during erythropoiesis. Heatmaps show repartition of the protein copy numbers ranked according to their transcripts features (CAI, 5’ and 3’UTR structure and total length) in the rows and stages of differentiation in the columns. Each row represents 1/10 of the total scale of the corresponding feature. For example, at the Epp stage 94.5% of the protein cellular content is issued from transcripts with a very high CAI (Top 10 % of CAI). Ret: reticulocytes; Eep: erythrocyte-enriched population; Epp: erythrocyte-purified erythrocytes.