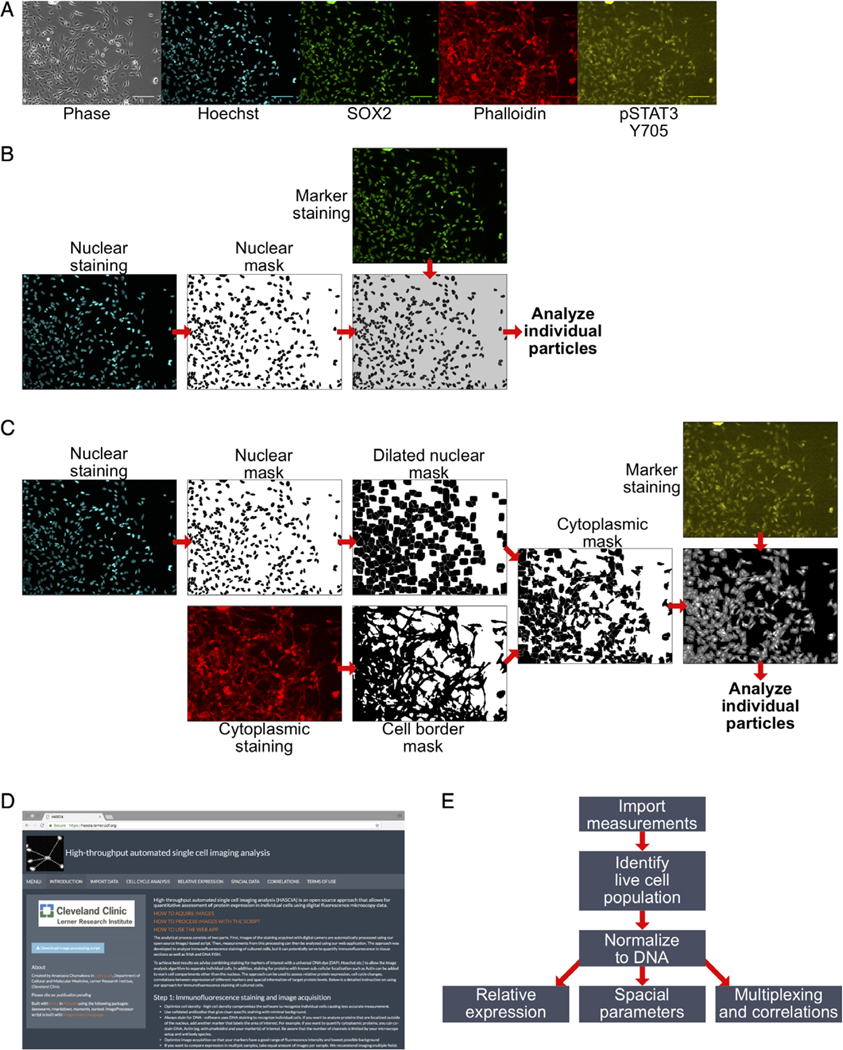
Figure 1.
High-throughput automated single cell imaging analysis (HASCIA) method description. (A) immunofluorescence images of an adherent GSC monolayer; scale bar, 100 μm; phase, phase contrast; Hoechst, DNA staining; SOX2, stem cell marker; phalloidin, B-actin staining; pSTAT3 Y705, phosphorylated Y705 of STAT3 staining. (B) Algorithm for nuclear staining analysis. Nuclear staining images are used to create nuclear masks and separating particles. The mask is further used for the delineation of nuclear region in nuclear marker staining. Individual particles are subsequently analyzed using the particle analysis feature of ImageJ. (C) Algorithm for cytoplasmic staining analysis. Nuclear mask is created as in B with subsequent dilation to fill perinuclear area. Cytoplasmic standard staining (in this case, actin, stained by Phalloidin-Alexa594) is used to create cell border outlines. Combined dilated nuclear mask and cell border mask yield cytoplasmic mask that is then used to delineate individual cellular cytoplasm on images of marker staining. (D) Screen shot of https://hascia.lerner.ccf.org. (E) Algorithm for HASCIA web-application data processing. After image processing with ImageJ scrip, measurements are imported into the app, based on DNA intensity profiles, live cell population for every specimen is outlined, and expression in specimens is calculated by applying the normalization of marker intensity to DNA intensity in each individual particle. The normalized data can further be used to compare expression between specimens, analyze local cell density, and perform multiplexing to investigate correlations between parameters. [Color figure can be viewed at wileyonlinelibrary.com]