Figure 3.
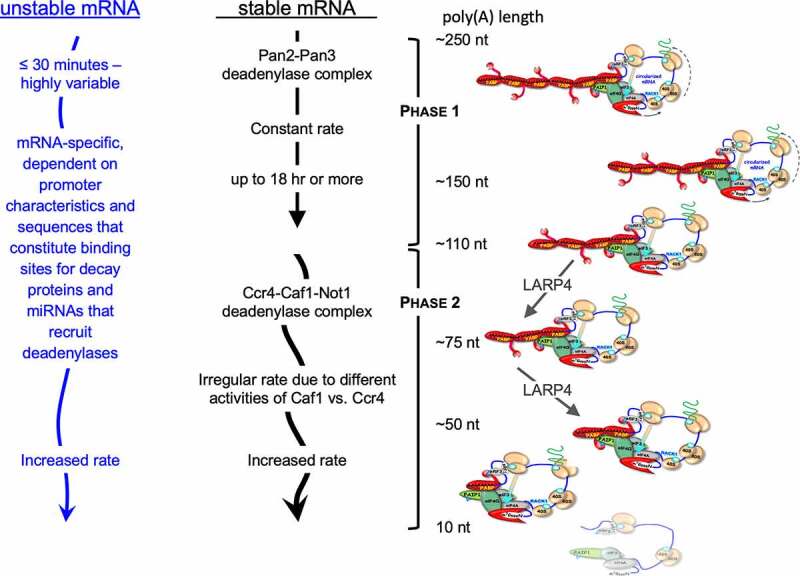
Schematic of model deadenylation pathways in mammalian cells. Model pathways for unstable mRNAs (left) and stable mRNAs (middle) derived from studies of reporter constructs and certain cellular mRNAs (see text). Classic stable mRNAs which have a paucity of destabilizing elements, follow a default pathway with biphasic deadenylation kinetics. The PAN2/3 initiates and carries out the first phase on nascent poly(A) tails of ~250 nucleotides with regular or constant rate depicted by straight downward arrow. After trimming to poly(A) lengths of 150–110 residues the mRNAs become preferred substrates of the CCR4-CAF1-NOT1 deadenylase complex CCR4-CNOT [.125,129] whose rate is irregular as depicted by the curved arrow. Unstable mRNAs, e.g., with AREs and/or miRNA binding sites in their 3ʹ UTR actively recruit deadenylases. Many mRNAs bear sequence elements or are engaged by other means that lead to active recruitment of the PAN2/3 and/or CCR4-NOT deadenylase complexes (see [134] and refs therein). The CCR4 and CAF1 deadenylase subunits of the CCR4-CNOT complex exhibit different types of deadenylase activity; CCR4 is active on poly(A) that is bound by PABP whereas CAF1 is active only on unbound substrate. Such differences as well as loss of cooperative interactions between PABP monomers and progressive instability on poly(A) contribute to the associated irregular and accelerating rates of deadenylation [88,129,143]. mRNA decay and translation rates are also coupled via codon content relative to tRNA activity [143]. Right: Schematized mRNP-PABP complexes with different poly(A) lengths. Diagonal green arrows depict poly(A) lengths representing those found by SM-PAT-seq profiling to be altered in cells in which LAReP4 was present or absent [149]. Two effects were observed; poly(A) length phasing of ribosomal protein-mRNAs suggested apparent impediment of conversion of mRNPs with presumed four PABPs to three PABPs in the presence of LARP4. Time-course profiling revealed apparent impediment of conversion of mRNPs with three PABPs to two PABPs in the presence of LARP4[149]
