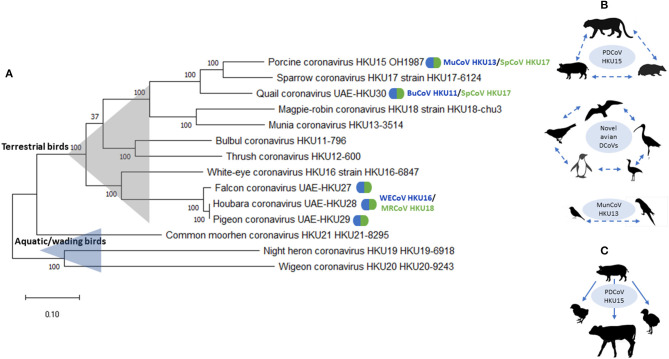
Figure 1.
(A) Phylogenetic analysis of complete genomic sequences of HKU11–HKU30 DCoVs. The evolutionary history was inferred by using the Maximum Likelihood method and General Time Reversible model. The tree with the highest log likelihood (−268,106.59) is shown. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 14 nucleotide sequences. Evolutionary analyses were conducted in MEGA X (47). Natural recombinant variants are  marked with. (B) Natural suggested host ranges are shown for PDCoV HKU15, MunCoV HKU13, and “novel” avian DCoVs. Dashed lines/arrows are used when the fact and direction of transmission cannot be confirmed due to the lack of complete genomic sequence and/or experimental evidence. (C) Experimental transmission and disease reproduction in chicks and poults using PDCoV HKU15.
marked with. (B) Natural suggested host ranges are shown for PDCoV HKU15, MunCoV HKU13, and “novel” avian DCoVs. Dashed lines/arrows are used when the fact and direction of transmission cannot be confirmed due to the lack of complete genomic sequence and/or experimental evidence. (C) Experimental transmission and disease reproduction in chicks and poults using PDCoV HKU15.