Figure 1.
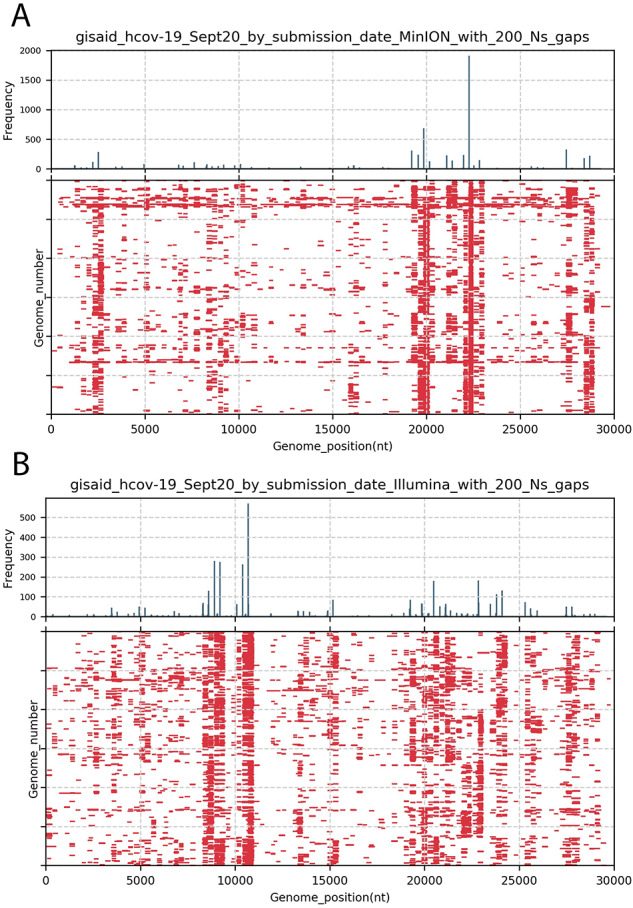
(updated graphics) Positions of 200 nt gaps across SARS-CoV-2 genomes listed as complete in GISAID. Genomes deposited in September 2020 (n = 38,228) were retrieved from GISAID, sorted by sequencing platform (MinION versus Illumina) and genomes with at least one instance of 200 N were collected. Panel A presents gaps in the first 3,000 MinION-generated genome sequences deposited that contained at least on 200 N motif. Gaps >= 200 nt in each genome are indicated with red bars. The upper panel histogram shows the frequency (in 30 nt bins) of gaps >=200 nt motifs by start position on genomes. Panel B is the same analysis of the first 3,000 Illumina-generated genome sequences in September 2020 that contained at least one 200 N motif.
