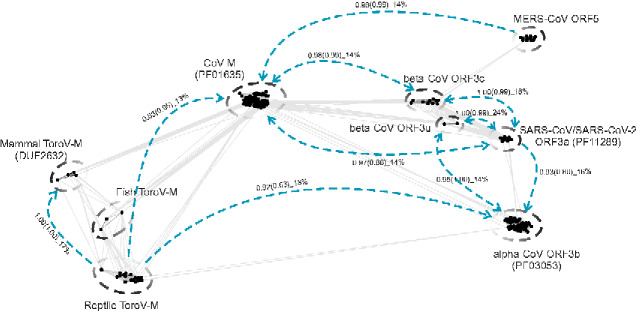
Figure 1.
Graphic two-dimensional representation of sequence similarity between the divergent M/ORF3 proteins of CoVs and ToroVs. The similarities were detected by both CLANS and HHsearch programs. Each node corresponds to a protein sequence. Straight lines indicate significant high-scoring segment pairs (HSPs) detected by all-against-all BLASTP searches with scoring matrix BLOSUM45 and e-value cutoff of 0.02. The graph was generated by CLANS, which uses the Fruchterman and Reingold graph drawing algorithm (Frickey and Lupas 2004). The gapped circles indicate individual protein families. Blue dashed lines indicate the remote relationship between families detected by the HHsearch program where the arrow indicates the search direction. The probabilities of the HHsearch comparisons with full-length protein sequences and C-terminal cytoplasmic β-sandwiches (in brackets) are indicated, followed by the percentage sequence identity.