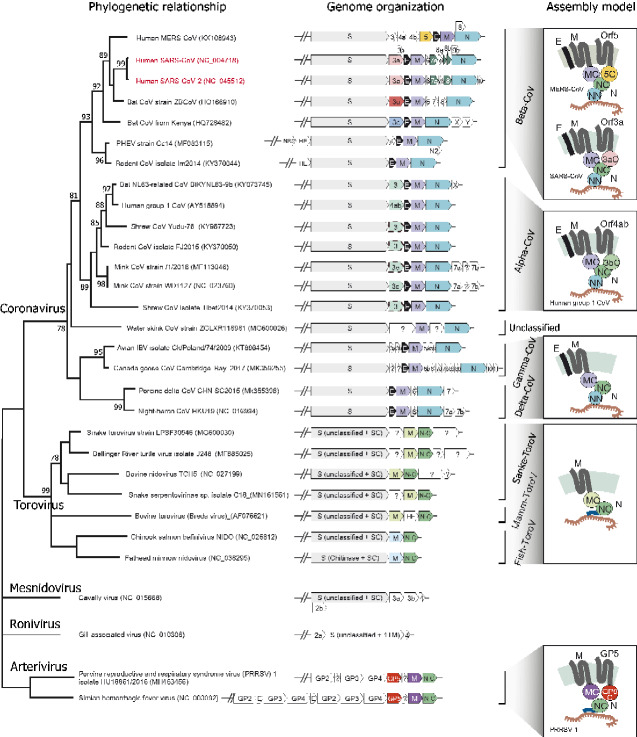
Figure 6.
Genomic organization of representative CoVs, ToroVs and other related nidoviruses. The detailed species tree of the CoV-ToroV clade was adapted from the Fig. 4. The genomic regions encoding for structural and accessory proteins are illustrated right to the terminal nodes of the phylogenetic tree. The gene names are shown according to their NCBI genome annotation, but the color of the gene box corresponds to different protein families: all CoV Spike proteins in grey, SARS-CoV/SARS-CoV-2 ORF3a in pink, beta-CoV ORF3u in red, MERS-CoV ORF5 in yellow, beta-CoV ORF3c in sky-blue, alpha-CoV-229E ORF3b in pale-green, typical CoV-M proteins in light-purple, reptile and mammal ToroV M protein in light-green, fish ToroV M proteins in light-blue, envelope (E) protein in black, SARS-CoV ORF7a-Ig and ORF8-Ig in dark-green, nucleocapsid (N) protein in blue-green, nucleocapsid (N) protein C-terminal dimerization domain in green, Arterivirus ORF3-like GP5 protein in vermilion and Arterivirus M protein in purple. Several previously unannotated alpha-CoV ORF3b genes are recovered and highlighted in dashed boxes. The predicted assembly models (without Spike) are shown on the right side of genomic organization. Protein abbreviations used in the model are as follows: MC: M protein C-terminal β-sandwich domain; NN: N protein N-terminal RBD domain; NC: N protein C-terminal dimerization domain; SC: Spike protein C-terminal domain; 5C: MERS-ORF5 C-terminal β-sandwich domain; 3aC: SARS-CoV/SARS-CoV-2 ORF3a C-terminal β-sandwich domain; 3bC: alpha-CoV-229E ORF3b C-terminal β-sandwich domain; GP5C: Arterivirus GP5 protein C-terminal β-sandwich domain.