FIGURE 1.
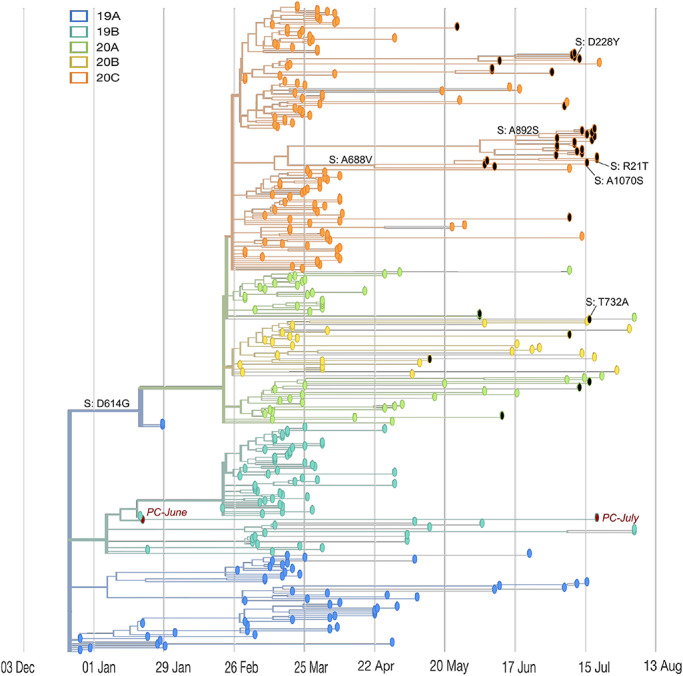
Genomic and epidemiological diversity of 59MDW SARS-CoV-2 sequences. The SARS-CoV-2 phylogeny was constructed in a local version of Nextstrain (Auspice.us) and shows the evolutionary relationship of 59MDW SARS-CoV-2 genomes (black dots) and a subsample of publicly available SARSCoV-2 genomes collected worldwide (n = 301, colored by viral clade [see legend]). This phylogeny contextualizes mutation rates and transmission patterns of SARS-CoV-2 strains collected at JBSA/Lackland and can be viewed locally using Auspice.us and a provided .json file. The tree is a time-resolved display rooted relative to early samples collected in Wuhan, China, and site numbering and genome structure uses Wuhan-Hu-1/2019 as reference. Positive control samples are shown in red and mutations in the spike protein are indicated in 59MDW samples.
