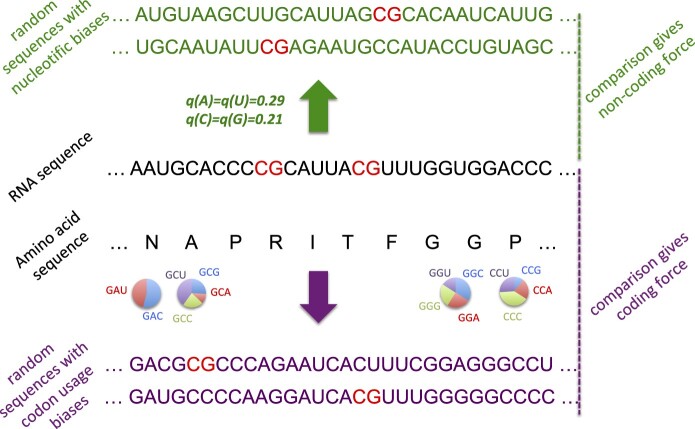
Fig. 1.
Schematic definitions of CpG noncoding and coding forces. The viral RNA sequence to be analyzed is shown in black, with CpG dinucleotide motifs in red. The force is computed by comparing the occurrences number of CpG with ensembles of random sequences fulfilling some of the constraints acting on the natural sequence, see Materials and Methods for details. Top, “noncoding” framework, in green: sequences of the same lengths can be generated by randomly drawing nucleotides according to prescribed frequencies, here taken from the human genome. Bottom: When the sequence under consideration codes for a protein (sequence of amino acids in black letters), random sequences (violet letters) can be generated in a “coding” framework as follows. For each amino acid, a licit codon is randomly chosen according to prescribed codon usage (here, computed from the coding regions in the human genome). Notice that the above computational frameworks are not restricted to CpG and can be applied to other dinucleotidic motifs.