Abstract
Coronavirus disease 2019 (COVID-19) outcomes are linked to host immune responses and may be affected by antiviral therapy. We investigated antibody and cytokine responses in ACTT-1 study participants enrolled at our center. We studied serum specimens from 19 hospitalized adults with COVID-19 randomized to treatment with remdesivir or placebo. We assessed severe acute respiratory syndrome coronavirus 2 antibody responses and identified cytokine signatures, using hierarchical clustering. We identified no clear immunologic trends attributable to remdesivir treatment. Seven participants were initially seronegative at study enrollment, and all 4 deaths occurred in this group with more recent symptom onset. We identified 3 dominant cytokine signatures, demonstrating different disease trajectories.
Keywords: COVID-19, remdesivir, ACTT-1, neutralization, serology, cytokine, trajectory
Among participants in the ACTT-1 study of remdesivir treatment of coronavirus disease 2019, we performed immunologic profiling. Initial absence of serum antibody activity on study enrollment was associated with mortality rate, with longitudinal disease trajectories characterized by serum cytokine signature.
The Adaptive COVID-19 Treatment Trial-1 (ACTT-1) clinical trial of remdesivir was the first double-blind, randomized, placebo-controlled trial in the United States to find a beneficial effect of antiviral therapy for coronavirus disease 2019 (COVID-19), shortening time to recovery in hospitalized adults [1]. The interplay between antiviral therapies like remdesivir and host immune responses to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has not been explored. Results of serologic assays quantitating both antibody binding and functional neutralization activity against SARS-CoV-2 are positively correlated with disease severity [2, 3]. However, both rely on the timely development of pathogen-specific antibodies, which may be affected by inhibition of viral replication [2, 4].
Several studies have investigated whether humoral responses may serve as biomarkers of disease outcome with mixed results [5, 6]. Similarly the impact of antiviral therapy on cytokine responses is unknown. While others have identified cytokine signatures associated with disease trajectory, severity, and survival [7–9], one hallmark of COVID-19 is the high degree of interpersonal variability in immune parameters, consistent with the wide spectrum of disease manifestations and severity. We therefore aimed to define humoral immune responses and cytokine trajectories within our institutional ACTT-1 study cohort.
Our analyses suggest that absence of humoral immunity at the time of study enrollment was associated with a more aggressive immunologic trajectory and worse survival. Hierarchical clustering of multiplex serum cytokine concentrations over time revealed distinct immune signatures, confirming both the high degree of heterogeneity among patients with COVID-19 but also demonstrating strong consistency in immune response over time within individuals.
METHODS
University of Minnesota ACTT1 Study Enrollment
Of 1063 participants enrolled in ACTT-1, 31 were at the University of Minnesota Medical Center and Bethesda Hospital, MHealth Fairview Medical Center. Our site adhered to the ACTT-1 study protocol for consent and enrollment of participants [1] and was overseen by a central institutional review board at the University of Nebraska. Hospitalized individuals ≥18 years old with symptoms consistent with COVID-19 and polymerase chain reaction–confirmed SARS-CoV-2 infection were included. All had either radiographic pulmonary infiltrate or hypoxemia (oxygen saturation ≤94% on room air). Participants were randomized to receive a 10-day treatment course of either remdesivir or placebo while hospitalized. Of 31 participants, 19 consented to collection of serum samples. Participant characteristics are in Supplementary Table 1.
Serum Collection
Serum was collected and stored at −80°C for batch analysis. Pretreatment (day 1) specimens were collected for all 19 participants, posttreatment (day 11 or 15) specimens were collected for 13 participants, and final (day 29) specimens were collected for 7 participants.
Serology
Anti–SARS-CoV-2 serologic status was determined using enzyme-linked immunosorbent assay (ELISA) kits from RayBiotech using recombinant SARS-CoV-2 nucleocapsid (N) or spike (S) receptor binding domain (RBD) antigens and detecting human immunoglobulin (Ig) G binding these antigens. A standard serum dilution of 1:100 was used for analysis of all samples. A seropositivity cutoff was chosen as the lowest optical density observed for each antigen for all participants at the posttreatment time point.
Live SARS-CoV-2 Virus Neutralization Assay
Neutralization assays were performed using an infectious clone of live SARS-CoV-2 expressing mNeonGreen fluorescent protein [10–12]. Vero E6 cells were seeded in a 96-well Black/Clear Flat Bottom TC-treated plate (Falcon). Serum samples were 2-fold serially diluted (from 1:20 to 1:5120) and incubated with mNeonGreen SARS-CoV-2 at 37°C for 1 hour, before infection at multiplicity of infection of 0.1 plaque-forming units per cell. After incubation at 37°C for 24–26 hours, cells were fixed in 4% paraformaldehyde, and fluorescence signal was detected on a BioTek Synergy H1 Hybrid Multi-Mode Reader (excitation, 488 nm; emission, 517 nm). A nonlinear regression method was used to calculate the serum titer that neutralized 50% of mNeonGreen fluorescence (NT50) using Prism 8 (GraphPad). Each serum sample was tested in duplicate. Neutralization curves for all samples tested are presented in Supplementary Figure 1.
Serum Cytokine Profiling
A commercial 48-plex human cytokine kit was purchased from EMD Millipore and run on a Luminex instrument (Bioplex 200) at the Cytokine Reference Laboratory at the University of Minnesota. Serum samples were analyzed in duplicate, and cytokine concentration values were interpolated from 5-parameter fitted standard curves. Two cytokines (interleukin 3 and macrophage inflammatory protein 1a) were undetectable across all samples tested and therefore excluded from all analyses. Hierarchical clustering and heat map generation were accomplished using the ComplexHeatmap package in R software, version 4.0.4. Each cytokine concentration was scaled and centered. Samples were initially clustered using an unsupervised hierarchical method, which identified 3 dominant immune signatures. Based on this analysis, final sample clustering was performed after manual assignment of each sample to 1 of these 3 clusters. A k-means algorithm was used to divide cytokines into 2 groups to identify early versus late features of infection.
RESULTS
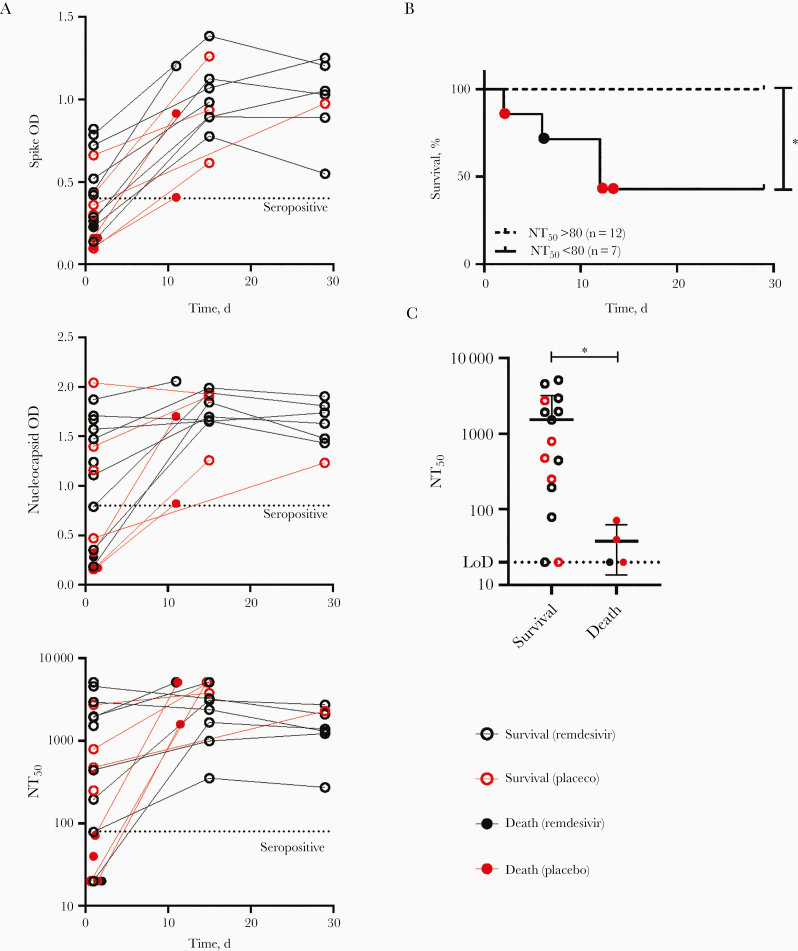
To longitudinally assess antibody responses, we performed ELISAs for nucleocapsid (N) and spike (S) RBD–specific IgG, as well as live SARS-CoV-2 neutralization assays, calculating the NT50 of viral infection. We found a broad range of initial serologic values, with some individuals presenting as seropositive on day 1 of enrollment, while others were seronegative (Figure 1A). We observed no clear association between treatment arm and serologic outcomes. Seven participants were initially seronegative by neutralization assay (NT50 <80), and 12 were seropositive (NT50 >80). All participants assessed at the posttreatment time points were seropositive by all assays (Figure 1A). Participants who were initially seronegative for both S RBD and N, or who were seropositive for N alone, had significantly lower neutralization activity than individuals that were initially seropositive for both S RBD and N (Supplementary Figure 2A). There was excellent agreement between ELISA and neutralization assay results for day 1 specimens, with strong positive associations between NT50 and both S RBD IgG (Spearman r = 0.9736 [Supplementary Figure 2B]) or N IgG (Spearman r = 0.8740 [not shown]).
Figure 1.
Association between severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibody responses and coronavirus disease 2019 mortality rate. A, Spike receptor binding domain and nucleocapsid immunoglobulin G levels for each participant were measured longitudinally, using optical density (OD) readout of enzyme-linked immunosorbent assays for a standard serum dilution. Neutralization activity was determined using a live SARS-CoV-2–mNeonGreen (mNG) reporter assay, reported as the serum titer preventing 50% of infection (NT50). Dotted line represents designated seropositivity threshold for each assay. B, Kaplan-Meier survival curves for patients with initial NT50 values >80 or <80 (P = .003; Mantel-Cox test). C, Dot plots comparing NT50 values on day 1 among participants grouped by 29-day survival outcome. Dotted line represents limit of detection (LoD; 1:20) of neutralization assay. Statistical analysis was conducted with Mann-Whitney U test; P = .01
Of 19 participants, all 4 who did not survive were seronegative for SARS-CoV-2 (S RBD and N IgG negative; NT50 < 80) at the time of enrollment. The median survival duration for the seronegative group was 12 days, versus 29 days for the seropositive group (P = .003; Figure 1B and Supplementary Table 2), and the median duration of hospitalization for the seronegative group was 34 days, versus 14.5 days in the seropositive group (P = .03; Supplementary Table 2). Concordantly, there was a significant difference in neutralization activity between the survivors and those who died (Figure 1C), with a median NT50 value among survivors of 1535 compared with 37.81 in the deceased group (Supplementary Table 2). There was no association between initial seropositivity and other patient characteristics, including sex, age, or study arm; however, the seronegative group was enriched for participants with a more recent onset of symptoms, compared with the seropositive group (median time since onset, 5 vs 9.5 days; Supplementary Table 2). These findings suggest that individuals who are seronegative at the time of initial hospital evaluation for COVID-19 may represent a subgroup at higher risk of poor outcome, owing to either a delayed onset of humoral immune responses or a more aggressive disease trajectory.
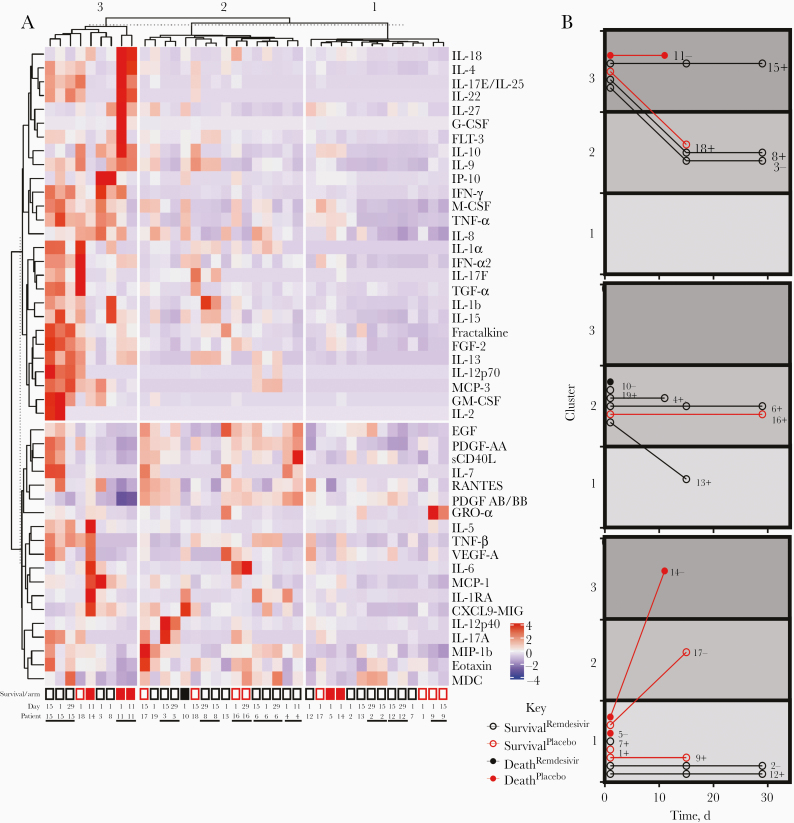
To further explore the relationship between cytokine profile and patient characteristics, we performed unsupervised hierarchical clustering of 46 cytokines from 39 serum specimens longitudinally collected from our 19 participants. Specimens clustered into 3 cytokine signatures characterized by high (cluster 3), moderate (cluster 2), and mild (cluster 1) overall cytokine expression (Figure 2A). These data are broadly similar to the cytokine signatures identified by others, with the hyperinflammatory cluster 3 comparing closely with clusters representing severe disease, characterized by the high expression of tumor necrosis factor a, interleukin 6, and macrophage colony-stimulating factor [9]. Cluster 1 was consistent with cytokine signatures of either early or moderate disease and recovery. Samples from the 4 individuals who did not survive were found in each of the clusters.
Figure 2.
Multiplex serum cytokine profiling reveals patient-specific signatures and inflammatory clusters consistent with more severe disease. A, Serum samples (columns) were subjected to unsupervised hierarchical clustering based on multiplex cytokine concentrations. Cytokine (rows) groupings were assigned based on a k-means set at 2. Participant identifier (ID) and serum collection study date are presented for each column, with neighboring samples from the same individual distinguished by horizontal black bars. Open boxes represent participants who survived, and closed boxes, participants who died. Black boxes represent participants who received remdesivir treatment, and red boxes, those who received placebo. B, Disease trajectories for each participant, represented by cytokine cluster assignment of each serum specimen collected over time. Participants are presented in 3 different time courses, based on the cytokine cluster of the day 1 serum specimen. Participants IDs and initial neutralization designation are presented next to each time course (plus signs represent serum titer preventing 50% of infection [NT50], >80; minus signs, NT50, <80). Abbreviations: EGF, epidermal growth factor; FGF, fibroblast growth factor; FLT, fms-like tyrosine kinase; G-CSF, granulocyte colony-stimulating factor; GM-CSF, granulocyte-macrophage colony-stimulating factor; GRO, growth-related oncogene; IFN, interferon; IL-1b (etc), interleukin 1b (etc); IP, interferon gamma-induced protein 10; MCP, monocyte chemoattractant protein; MDC, macrophage-derived chemokine; MIG, monokine induced by gamma interferon; MIP, macrophage inflammatory protein; PDGF, platelet-derived growth factor; RANTES, regulated on activation of normal T cells expressed and secreted; sCD40L, soluble cluster of differentiation antigen 40 ligand; TGF, transforming growth factor; TNF, tumor necrosis factor; VEGF, vascular endothelial growth factor.
Among pretreatment or posttreatment samples, we also found no association between cytokine cluster and treatment arm. Notably, we observed a close relationship in cytokine signatures among samples taken from the same individual. Indeed, of the 14 individuals with >1 sample collected in our data set, 9 had multiple samples that clustered together despite their collection being temporally separated by 10–28 days (Figure 2A). These results likely reflect the observed heterogenous spectrum of COVID-19 disease severity but reveal immunologic consistency over time within individuals.
Because our cytokine data set revealed a high degree of individuality among participants, we finally focused on each participant’s disease trajectory over time, as marked by progression between cytokine clusters. By comparing all participants with initial samples localized to the same cluster, we identified 3 major trajectories for movement between clusters: stable, increasing, and decreasing (Figure 2B). Of the 14 participants with >1 sample collected, only 2 (participants 017 and 014) demonstrated an increasing trajectory over time, both starting from the mild disease cluster 1. Both of these individuals were initially seronegative and were enrolled after a brief duration of illness (4 and 5 days), suggestive of an aggressive disease trajectory. Eight participants showed a stable progression over time. Two (participants 015 and 011), both initially seronegative, remained in the most severe disease cluster. All participants with stable disease trajectories remaining in less severe clusters 1 or 2 survived the infection, and only 1 of these was seronegative on day 1.
These limited data suggest an association between the prompt initiation of pathogen-specific humoral immunity and a more stable, potentially less lethal disease course. Finally, 4 participants had decreasing disease trajectories, suggesting a resolving infection, and all of them survived the disease. The individuality of our cytokine data set as a whole suggests that, depending on a patient’s immunologic status and timing of sample collection, a cytokine signature at a single time point may be insufficient to determine patient trajectory and prognosis.
DISCUSSION
Our study of immunologic characteristics of 19 hospitalized participants with COVID-19 enrolled in the ACTT-1 trial revealed several key insights into the pathogenesis of this heterogenous disease. Within our small cohort, we observed that the absence of serum neutralization at enrollment was associated with worse patient outcomes, including worse 29-day survival and longer hospitalizations among those surviving. These data suggest that initial antibody levels in hospitalized patients could provide valuable prognostic data for stratifying patients according to risk. While neutralization assays with live SARS-CoV-2 are resource intensive, ELISA-based methods could provide a suitable alternative in acute illness, where the correlation between antibody binding and functional activity is strong [13, 14]. Furthermore, lower neutralization titer at enrollment was associated with a shorter time from symptom onset to enrollment, suggesting more rapid disease progression characterized by symptom progression that outpaces the initiation of adaptive immunity.
While larger studies have identified several cytokines as biomarkers of disease severity and outcome, in our smaller study, we did not identify any individual cytokines whose pretreatment or posttreatment values were strongly predictive of mortality rate (Supplementary Tables 3 and 4). This difference may reflect the limited power of our small sample size to detect these differences, in the context of substantial disease heterogeneity. Initial platelet-derived growth factor AA/AB serum levels were significantly higher among individuals who survived (Supplementary Table 3) potentially identifying participants with more effective adaptive immunity at the time of enrollment and a relatively benign disease trajectory.
Although we did observe associations between mortality rate and both initial seronegativity and recency of symptom onset, we did not identify large differences in antibody or cytokine responses between remdesivir and placebo treatment groups. This may reflect the limited effect of initiating antiviral therapy on immune trajectories already in progress, or the limited power of our small data set to detect these differences. Yet, our cytokine analyses add to the growing appreciation of the individuality of each patient’s response to SARS-CoV-2 infection and disease trajectory, which may have substantial impact on the universality of biomarker interpretations [8]. A larger-scale analysis of serology, neutralization, and cytokine signature in the multicenter ACTT-1 data set could contribute more information on the impact of remdesivir on host immune responses and individual COVID-19 disease trajectories.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Allison Weyer, University of Minnesota (UMN) ACTT-1 research coordinators Mary Bailey, Jamie Heisdorffer, Kyong Yun, Mia Larson, Coley Landvik, Dean Krueger, Michelle Pitt, Mandi DeGrote, and Tommy Goodwin, and the UMN Cytokine Reference Laboratory.
Financial support. This work was supported by the National Institutes of Health (grants T32AI055433 to J. M. T., T32HL774126 to W. E. M., and 1R01AI153602 to V. D. M.) and UMN Department of Medicine funding (to T. D. B.).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Beigel JH, Tomashek KM, Dodd LE, et al. . Remdesivir for the treatment of Covid-19—final report. N Engl J Med 2020; 383:1813–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Long QX, Liu BZ, Deng HJ, et al. . Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 2020; 26:845–8. [DOI] [PubMed] [Google Scholar]
- 3. Krammer F, Simon V. Serology assays to manage COVID-19. Science 2020; 368:1060–1. [DOI] [PubMed] [Google Scholar]
- 4. Premkumar L, Segovia-Chumbez B, Jadi R, et al. . The receptor-binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci Immunol 2020; 5:eabc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen Y, Zuiani A, Fischinger S, et al. . Quick COVID-19 healers sustain anti-SARS-CoV-2 antibody production. Cell 2020; 183:1496–507.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takahashi T, Ellingson MK, Wong P, et al. ; Yale IMPACT Research Team . Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature 2020; 588:315–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lucas C, Wong P, Klein J, et al. ; Yale IMPACT Team . Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 2020; 584:463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Laing AG, Lorenc A, Del Molino Del Barrio I, et al. . A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat Med 2020; 26:1623–35. [DOI] [PubMed] [Google Scholar]
- 9. Valle DMD, Kim-Schulze S, Huang HH, et al. . An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med 2020; 26:1636–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gniadek TJ, Thiede JM, Matchett WE, et al. . SARS-CoV-2 neutralization and serology testing of COVID-19 convalescent plasma from donors with nonsevere disease. Transfusion 2021; 61:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xie X, Muruato A, Lokugamage KG, et al. . An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe 2020; 27:841–8.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Muruato AE, Fontes-Garfias CR, Ren P, et al. . A high-throughput neutralizing antibody assay for COVID-19 diagnosis and vaccine evaluation. Nat Commun 2020; 11:4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chi X, Yan R, Zhang J, et al. . A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 2020; 369:650–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ren L, Fan G, Wu W, et al. . Antibody responses and clinical outcomes in adults hospitalized with severe coronavirus disease 2019 (COVID-19): a post hoc analysis of LOTUS China trial. Clin Infect Dis 2020. doi: 10.1093/cid/ciaa1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.