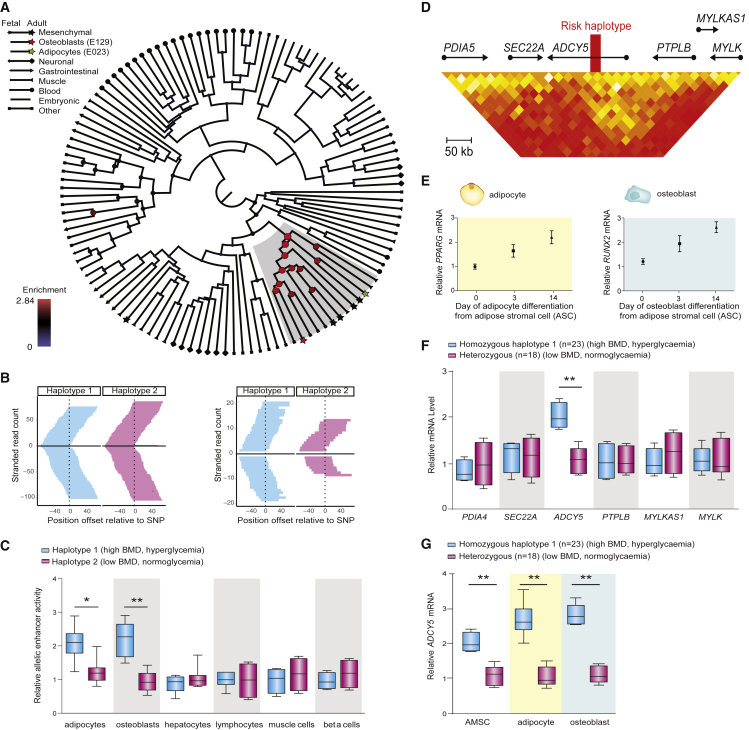
Figure 2.
ADCY5 expression differences between haplotypes in adipocytes and osteoblasts
(A) H3K27me3 enrichment at 3q21.1 across multiple cell types. Linkage cluster tree displays node sizes and colors proportional to the H3K27me3 enrichment. Mesenchymal cells, adipocytes, and osteoblasts are highlighted by colored stars.
(B) Allelic imbalance analysis of the 3p21.1 risk locus for H3K27me3 and chromatin accessibility by ATAC-seq in heterozygous AMSCs comparing haplotype 1 (blue) and haplotype 2 (pink). Each panel depicts H3K27me3 ChIP-seq and ATAC-seq read counts separated by strand above/below the midpoint.
(C) Haplotype-specific luciferase assays for 10-kb fragments containing 10 candidate regulatory SNPs from each haplotype in tight LD with rs2124500 (R2 > 0.9) in adipocytes, osteoblasts, hepatocytes, lymphocytes, differentiated muscle cells, and pancreatic beta cells. Mean ± SD, ∗p < 0.05, ∗∗p < 0.01.
(D) Genome-wide higher order chromatin interactions for the ADCY5 locus analyzed by Hi-C assays in human embryonic stem-cell-derived MSCs (Dixon et al., 2015).
(E) Quantification of adipocyte (yellow) and osteoblast (blue) differentiation marker gene expression from qPCR analysis across the course of differentiation. Three biological replicates, mean ± SD.
(F) Quantification of 3p21.1 risk haplotype-dependent candidate target gene expression from qPCR analysis in AMSC-derived osteoblasts from n = 18 haplotypes 1/2 carriers and n = 23 haplotypes 1/1 carriers. Mean ± SD, ∗∗p < 0.01.
(G) Quantification of 3p21.1 risk haplotype-dependent ADCY5 target gene expression from qPCR analysis in AMSCs, AMSC-derived osteoblasts and AMSC-derived adipocytes from n = 18 haplotypes 1/2 carriers and n = 23 haplotypes 1/1 carriers. Mean ±SD, ∗∗p < 0.01.