Figure 1.
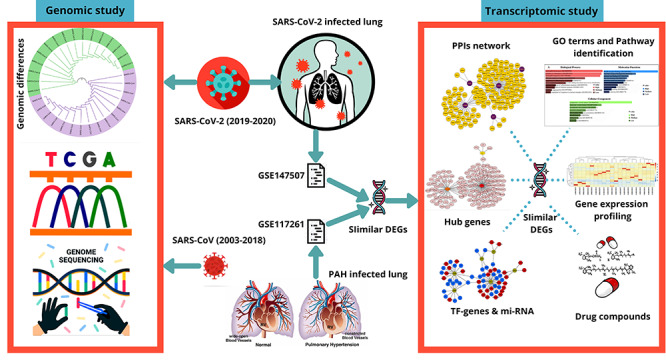
The workflow of current analysis. Genomic differences between SARS-CoV and SARS-CoV-2 are visualized through a phylogenetic analysis. Two datasets GSE147507 and GSE117261 are collected according to SARS-CoV-2 infection in human lung epithelial cells and PAH lung, respectively. Differentially expressed genes (DEGs) were identified using R programming language and similar DEGs were identified from total DEGs of both the datasets. Corresponding similar DEGs were used to perform transcriptomic analyses. The gene expression profiling was performed for both the datasets, and gene ontology (GO) terms, cell informative pathways, PPIs network, hub gene identification and TF–miRNA-based analyses were performed. According to the corresponding similar DEGs, drug compounds were predicted.
