Fig. 1. Phylogenetic distribution of d-aa synthetic and degradative enzymes.
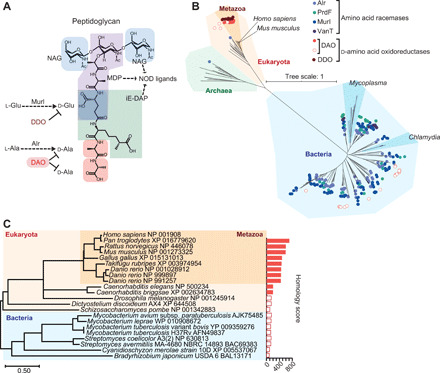
(A) The basic pattern of peptidoglycan (Gram-negative bacteria) shows incorporation of d-aa and innate ligands in the structure. NAG, N-acetylglucosamine; MDP, muramyl dipeptide; iE-DAP, γ-d-glutamyl-meso-diaminopimelic acid; NOD, nucleotide-binding oligomerization domain protein. (B) Amino acid racemases and d-aa oxidoreductases in the tree of life are shown. Circles indicate gene presence. Hollow circles indicate DAO genes with low homology (homology score < 200) to that of Homo sapiens. (C) A phylogenetic tree of DAO was analyzed using amino acid sequences of DAO. Bar graph represents BLAST homology scores. Hollow bars denote homology scores <200.
