Fig. 3. DAO-sensitive microbes promote IgA class switching.
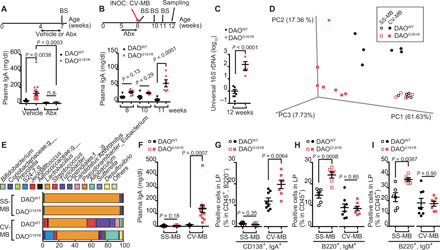
(A to I) DAOWT/WT and DAOG181R/G181R mice were treated with antibiotics or vehicle (A) (n = 9, 21, 12, and 9 mice), were treated with antibiotics followed by inoculation with CV-MB (B and C) (n = 7 and 6 mice), or were cross-fostered by dams colonized with SS-MB or CV-MB (D to I). BS, blood sampling; INOC, inoculation; Abx, antibiotics. (A, B, and F) Plasma IgA was measured with enzyme-linked immunosorbent assay (ELISA). (C) Amounts of SI microbiota were assessed with quantitative polymerase chain reaction (qPCR) of 16S ribosomal DNA (rDNA). (D) This plot indicates the principal coordinate analysis of unweighted UniFrac distances of SI microbiota that is detected by 16S rDNA metagenomic sequence. (E) Relative abundance of SI microbes is shown (average of n = 5 or 6 individuals per group). (G to I) Percentage of IgA+-plasma cells (G) or naïve B cells (H and I) in CD45+ cells in SI LP from DAOWT/WT and DAOG181R/G181R mice with indicated gut microbiota (n = 6, 6, 8, and 7 mice). Error bars, means ± SEM. Significance was analyzed with one-way ANOVA (A), two-way ANOVA (B), two-tailed unpaired t tests (C and G to I), and t tests with Welch’s correction (F).
