Fig. 5. DAO regulates cytokines in the SI epithelium.
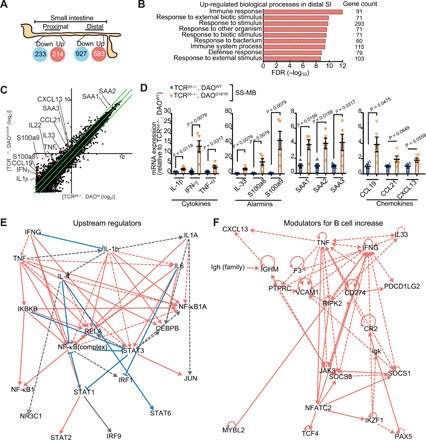
(A) Number of up-regulated or down-regulated genes in proximal and distal SI in TCRβ−/−,δ−/−/DAOG181R/G181R versus TCRβ−/−,δ−/−/DAOWT/WT mice colonized with SS-MB. (B) Functional classifications of significantly up-regulated genes in distal SI from TCRβ−/−,δ−/−/DAOG181R/G181R mice. FDR, false discovery rate. (C) Scatter plot showing the twofold or more up-regulated and down-regulated genes in log 2 scale. (D) mRNA expression of indicated cytokines, alarmins, serum amyloids, and chemokines in distal SI epithelium was analyzed by qPCR (n = 5 mice per group). (E) Top-scoring upstream regulators included several cytokines such as IFN-γ, IL-1β, IL-1α, IL-6, IL-4, and TNF, which were predicted to be extracellular upstream regulators. Solid line, leads to activation. Dashed line, leads to inhibition. Red line, findings consistent with state of downstream molecule. Blue, findings inconsistent with downstream. Gray, effect not predicted. (F) Genes associated with increased abundance of B cells suggest that TNF, IFN-γ, IL33, and CXCL13 are key regulators. Error bars, means ± SEM. Significance was analyzed with t test with Welch’s correction (D).
