Figure 1.
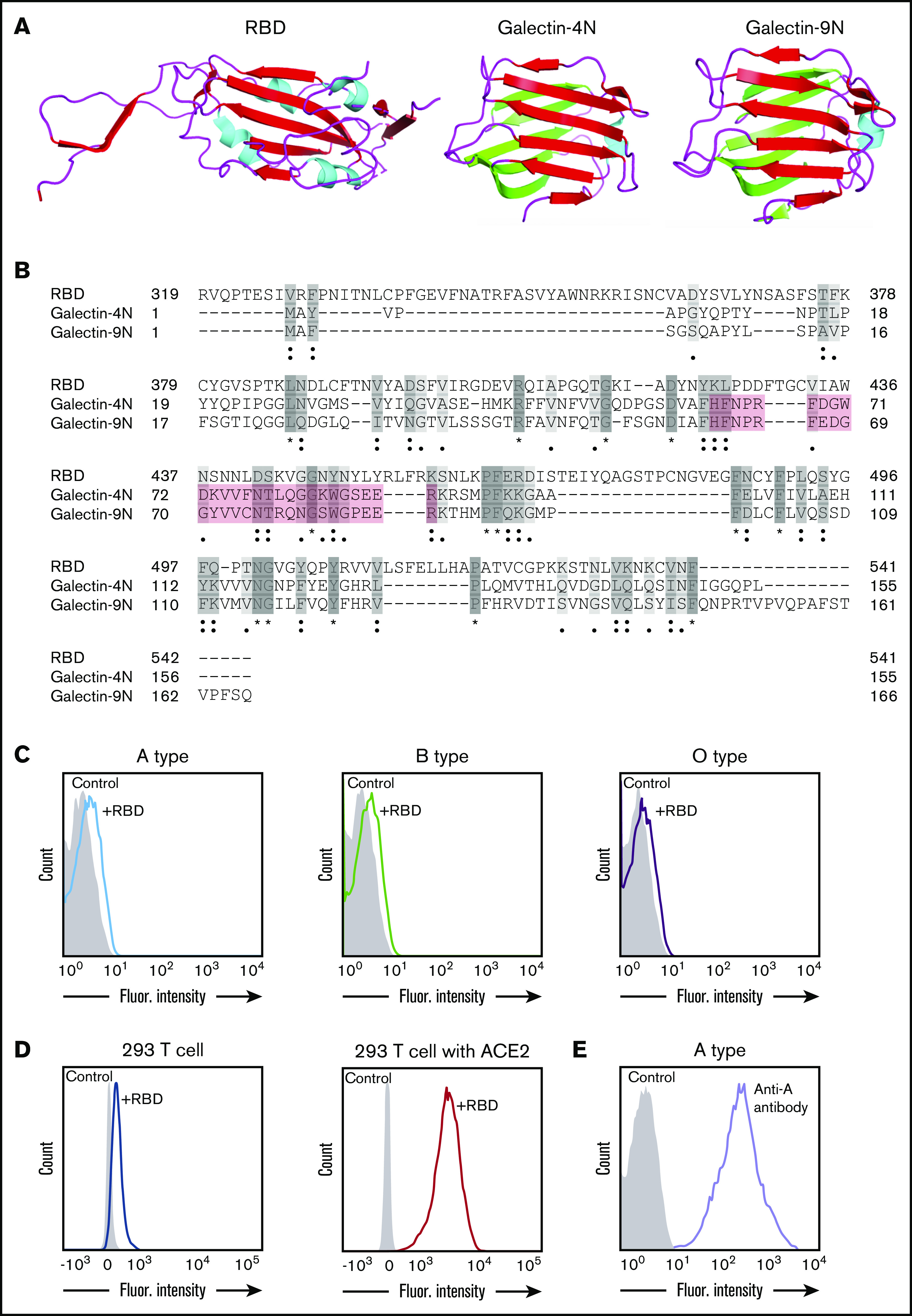
SARS-CoV-2 shares significant similarity to human galectins. (A) Structural representation of SARS-CoV-2 RBD (Protein Data Bank [PDB] ID: 6VXX), human galectin-4N-terminal domain (PDB: 5DUW), and human galectin-9N-terminal domain (PDB ID: 3LSD); β-sheet structures shown in red or green, α-helix shown in cyan. (B) Sequence alignment (Uniport align program) of SARS-CoV-2 RBD, human galectin-4N-terminal domain (galectin-4N) and human galectin-9 N-terminal domain (galectin-9N) indicating fully conserved residues (dark gray [*]), highly conserved residues (gray [:]), lowly conserved residues (light gray [.]); the galectin carbohydrate binding pocket is highlighted in light red. (C) Flow cytometric analysis of blood group A, B, or O type human RBCs after incubation with SARS-CoV-2 RBD for 1 hour followed by detection with anti-His antibody. (D) Flow cytometric analysis of HEK293 T cells alone or HEK293 T cells stably transduced to express ACE2 after a 1-hour incubation with the SARS-CoV-2 RBD and detection with anti-His antibody. (E) Flow cytometric examination of blood group A RBCs after incubation with anti-A antibody. Secondary antibody alone served as a control.
