Figure 2.
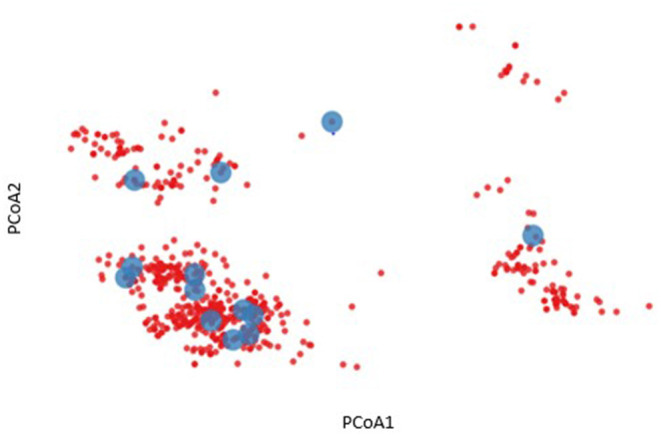
Two-dimensional representation of HLA diversity using Principal Component Analysis (PCoA). A HIV-1 Gag binding profile was predicted for every HLA allele using NetMHCpan and a set of transmitted founder sequences. The binding profile of each volunteer (red dot) was defined by taking the union of predicted binding for each of their HLA alleles. PCoA was performed using the pairwise similarity matrix of all volunteers, revealing distinct clusters of individuals. A subgroup of 13 volunteers were chosen to provide optimal coverage of the HLA binding profiles (blue dots).
