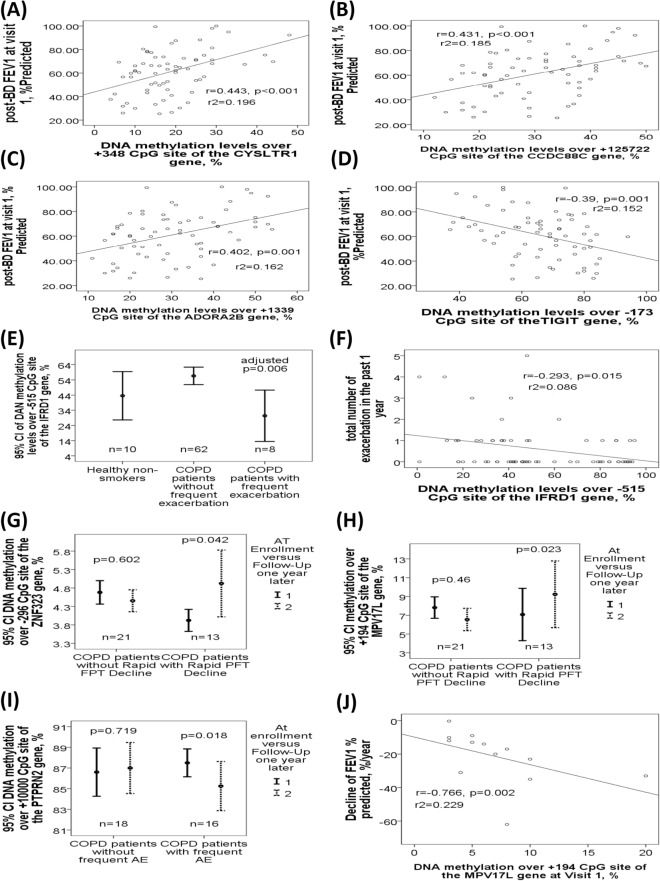
Figure 3.
Differential DNA methylation patterns of the IRFD1, TIGIT, CysLTR1, ADORA2B, CCDC88C, ZNF323, and MPV17L, and PTPRN2 genes at cross sectional or longitudinal levels in the validation cohort. (A) DNA methylation levels of the CYSLTR1 gene promoter region (+ 348 CpG site) were positively correlated with post-BD FEV1%predicted value. (B) DNA methylation levels of the CCDC88C gene body (+ 125,722 CpG site) were positively correlated with post-BD FEV1%predicted value. (C) DNA methylation levels of the ADORA2B gene body (+ 1339 CpG site) were positively correlated with post-BD FEV1%predicted value. (D) DNA methylation levels of the TIGIT gene promoter region (− 172 CpG site) were negatively correlated with post-BD FEV1%predicted value. (E) DNA methylation levels of the IFRD1 gene promoter region (− 515 CpG site) were decreased in all the COPD patients with frequent exacerbation versus those without frequent exacerbation, and (F) negatively correlated with the number of exacerbations in the past one year. (G) DNA methylation levels of the ZNF323 gene promoter region (− 296 CpG site) were elevated after 1-year follow-up in COPD patients with rapid lung function decline versus that at enrollment. (H) DNA methylation levels of the MPV17L gene (+ 194 CpG site) were elevated after 1-year follow-up in COPD patients with rapid lung function decline versus that at enrollment. (I) DNA methylation levels of the MPV17L gene (+ 194 CpG site) at visit 1 were negatively correlated with the difference in FEV1% predicted values between visit 2 and visit 1. (J) DNA methylation levels over + 10,015 CpG site of the PTPRN2 gene were reduced after 1-year follow-up in COPD patients with frequent exacerbation versus that at enrollment.