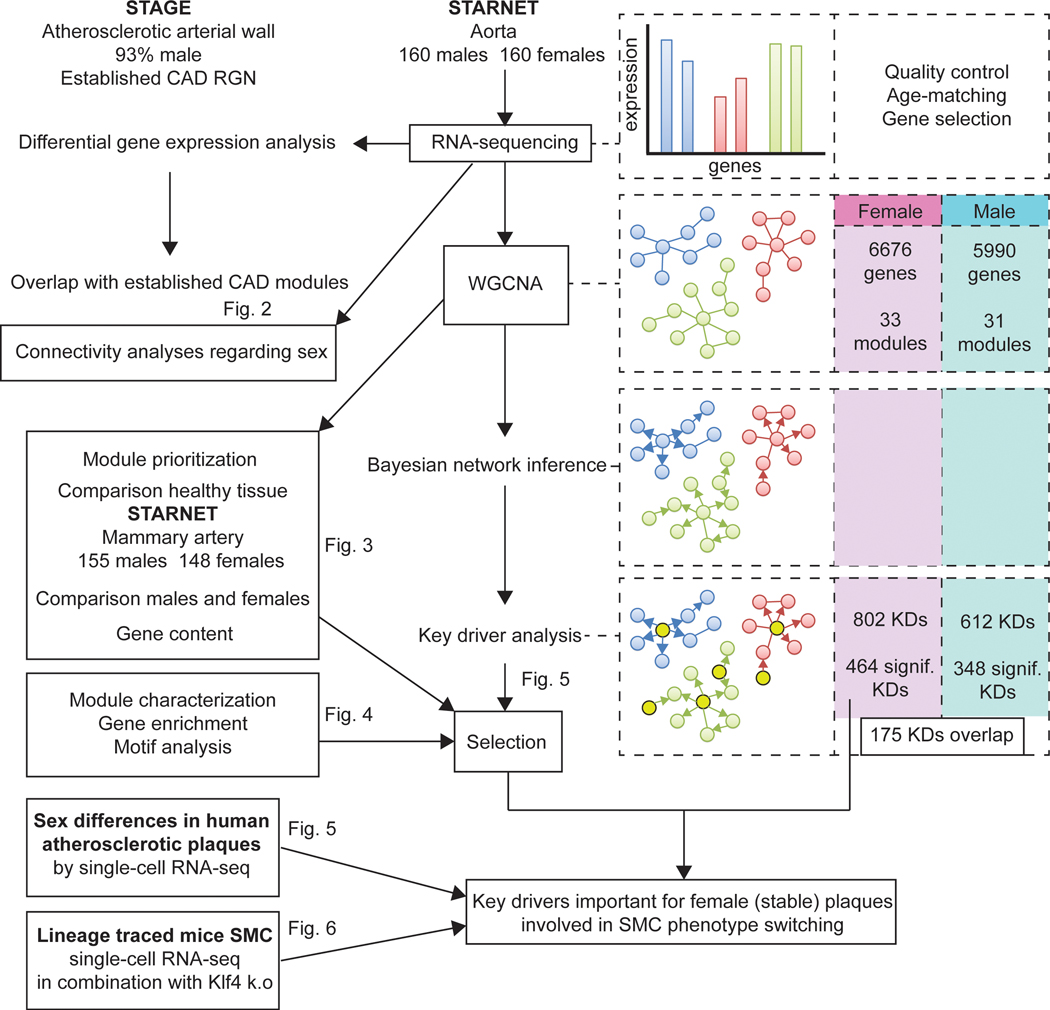
Figure 1. Workflow of sex-stratified GRN analyses.
The workflow of all analyses performed in this study are shown. We started by analysing previous determined CAD GRNs in STAGE (90%+ male) by using DEG data between the sexes from AOR in STARNET. To determine whether connectivity is affected by sex, we calculated gene connectivities in a multitude of populations in which we changed the proportion of females and males. Next, we generated sex-stratified GRNs by WGCNA, Bayesian network inference and key-driver analysis. The number of sex-stratified genes, modules, and key drivers are noted in the right boxes. We prioritized modules created by WGCNA for their importance in female CAD, by comparing how these modules behaved in healthy arterial tissue (MAM) and between the sexes. Further, we correlated the expression of the gene contents with clinical parameters of CAD, measured in STARNET, among which HDL, LDL, TG, glucose, and CRP levels, as well as Syntax score (a measure for the abundance and complexity of coronary atherosclerosis). After prioritization, we annotated the modules and analysed their gene content by gene enrichment analyses and motif analyses. We then selected the key drivers for the female CAD modules and determined their sex-specifc cellular expression by single-cell RNA-sequencing of atherosclerotic plaques. Lastly, we used lineage traced mouse SMC single-cell RNA-seq data to determine female key driver functionality. AOR = atherosclerotic aortic root, CAD = coronary artery disease, GRN = gene regulatory network, MAM = mammary artery, SMC = smooth muscle cell, WGCNA = weighted gene co-expression analysis