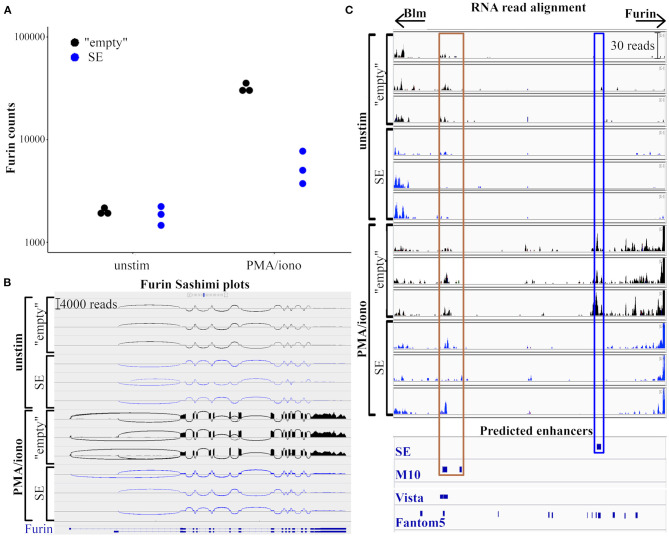
Figure 3.
Deletion of putative Furin super-enhancer does not affect the RNA splicing. RNA expression in “empty” and SE EL-4 cell clones was analyzed using RNA sequencing. (A) Dot plot represents the counts of RNA reads aligning to Furin gene in unstimulated or 6 h PMA/ionomycin stimulated clones. (B) Sashimi plots of Furin region (chr7: 80,389,000-80,406,000; mm10 assembly) generated with integrative genomics viewer (IGV). The heights of the bar diagrams are proportional to read counts aligning to the given region. Loops represents reads aligning to more exons, the thickness of the line refers to the number of bridging reads. (C) Read coverage of the genomic region between Furin and Blm genes (chr7: 80,405,000–80,455,000; mm10 assembly). The modified IGV view shows RNA read alignments in unstimulated or PMA/ionomycin-stimulated “empty” and SE cell clones, as well as predicted enhancer sites (SE: putative super-enhancer (deleted in SE clones), M10: Furin enhancer (48), Vista: enhancers from Vista enhancer browser (49), Fantom5: permissive enhancers according to Fantom5 enhancer atlas (50). Brown frame marks genomic areas with validated Furin enhancer activity, blue frame marks the putative super-enhancer.