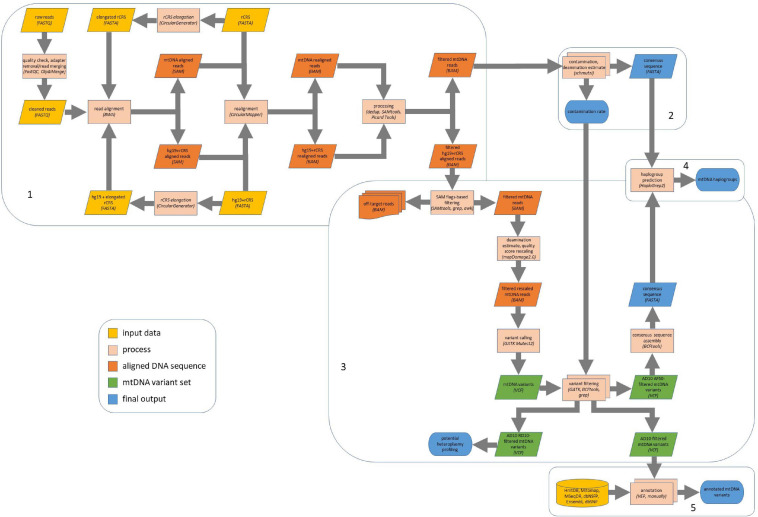
FIGURE 1.
Computational pipeline for ancient mitochondrial DNA (mtDNA) analysis. Our computational pipeline comprises five main steps: (1) read alignment and preprocessing and postprocessing; (2) contamination analysis by schmutzi and consensus sequence assembly; (3) variant calling by GATK Mutect2, variant filtering, and consensus sequence assembly; (4) haplogroup prediction; (5) variant annotation. The alignment required revised Cambridge Reference Sequence (rCRS) as reference sequence to get a suitable input to schmutzi, while we used mtDNA reads aligned onto the whole genome (hg19 with rCRS as a mitochondrial reference sequence) for variant calling. AD10, minimum variant allele depth = 10; AF50, minimum allele fraction = 50%; RD10, minimum reference allele depth = 10.