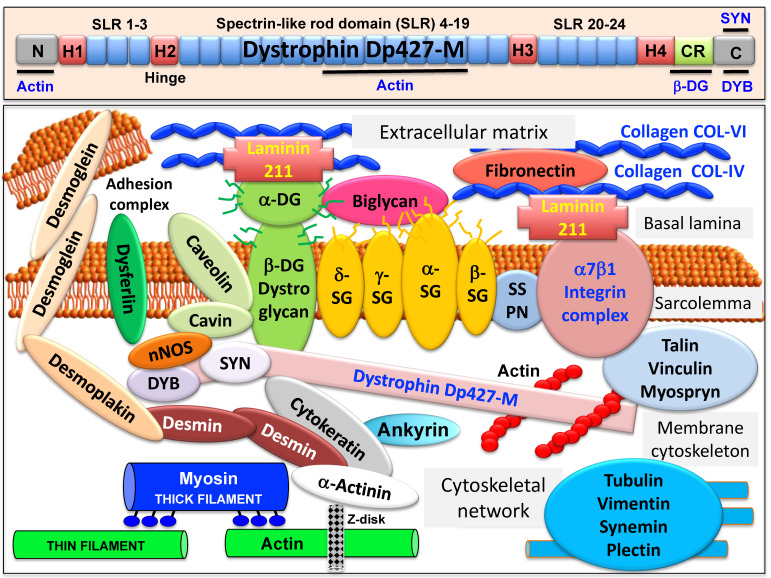
Figure 1.
Overview of the domain structure of dystrophin and the diverse interactions of the dystrophin–glycoprotein complex in skeletal muscle tissues. The upper panel shows a diagrammatic presentation of the main molecular domains of dystrophin isoform Dp427-M, including actin-binding sites at the N-terminus and central rod domain, proline-rich hinge regions (H1 to H4), spectrin-like rod (SLR) domains 1–3, 4–19 and 20–24, a cysteine-rich domain with binding sites for integral beta-dystroglycan (DG), the cysteine-rich domain (CR) and the C-terminus with binding sites for dystrobrevin (DYB) and syntrophin (SYN). The lower panel shows a model of the spatial configuration of the dystrophin complexome in skeletal muscle fibers. Shown is the dystrophin core complex consisting of the dystrophin isoform Dp427-M, dystroglycans (DG), sarcoglycans (SG), sarcospan (SSPN), syntrophins (SYN) and dystrobrevins (DYB), as well as the wider dystrophin-associated network that forms associations with the extracellular matrix, the sarcolemma, the cytoskeleton and the sarcomere.