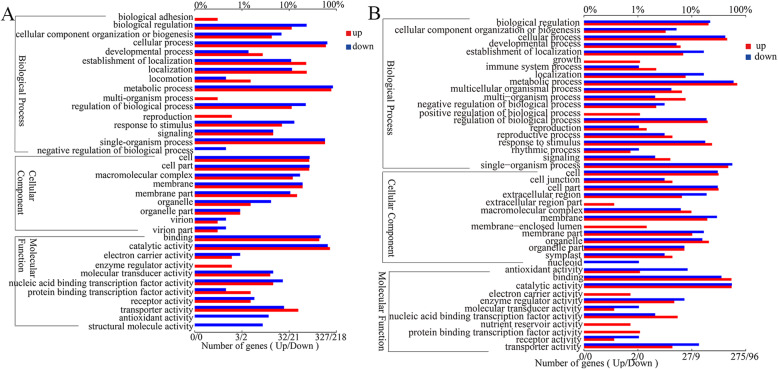
Fig. 4.
Enriched Gene Ontology (GO) terms distributed to the DEGs in P. polymyxa SC2 (a) and pepper (b).
DEGs from P. polymyxa SC2 interacting with pepper, as identified by RNA-Seq, were enriched based on the GO database. The abscissa below the figure indicates the number of genes annotated to a GO term. The upper abscissa indicates the proportion of the number of genes annotated to a GO term to the total number of all GO-annotated genes. (Genes and GO terms are many-to-many relationships; a gene can contain multiple GO term annotations, and a GO term can also correspond to multiple genes, not one-to-one relationships). Ordinates represent each detailed classification of GO. Three squares represent three secondary classifications of GO, respectively