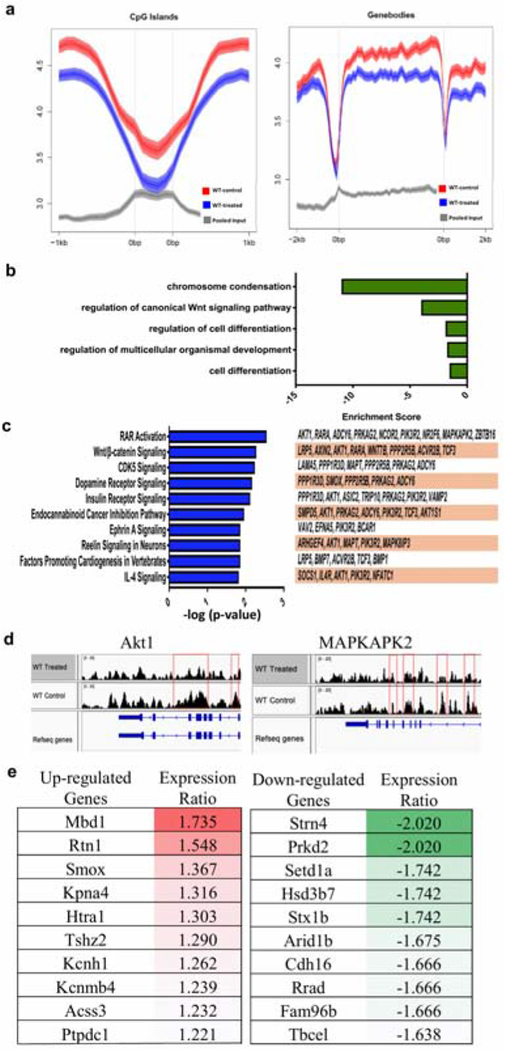
Figure 3. Genome-wide mapping of 5-hmC in epithelial samples from IMQ-treated skin VS untreated skin in mice.
(a) Data obtained from 2 biological replicates (n=2). Loss of 5-hmC is seen in both CpG islands and genebodies in IMQ-treated epidermis compared to untreated epidermis. (b) GO terms of downregulated pathways found in IMQ-treated skin. (c) Canonical pathway analysis of 377 differentially hydroxymethylated genes. The top 10 statistically significant canonical pathways identified by IPA shown here with genes identified by our study listed to the right. (d) hMeDIP results showing peaks correlating with 5-hmC levels at the promoter regions of Akt1 and MAPKAPK2 genes. (e) Top upregulated and downregulated genes identified in IMQ-treated lesional epidermis compared to non-lesional epidermis.