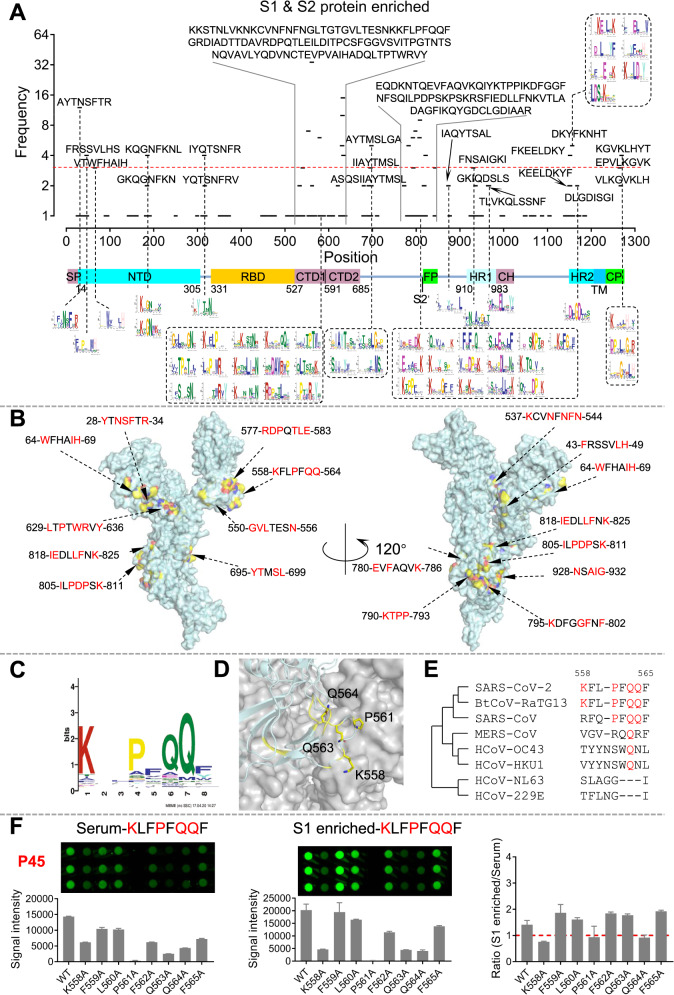
Fig. 1.
Significant epitopes on the S protein identified by AbMap and validated by peptide microarray. A The distribution of the sequence-matched epitopes on the S protein. B The distribution of the significant epitopes (frequency ≥ 3) on the 3D structure of the S protein monomer. Red amino acids represent key residues of epitopes. C A representative significant epitope. D The epitope was matched to the S protein, and critical residues were labeled yellow. E Homology analysis of the epitope among coronaviruses. F Validation of epitope identification procedures by a peptide microarray: comparison of sera and protein-enriched antibodies. Peptide 1, which corresponds to Epitope-S1-8 (KLFPFQQF), was selected as an example. The ratio of signal intensity (S1 enriched)/signal intensity (serum) for each residue was plotted (right)