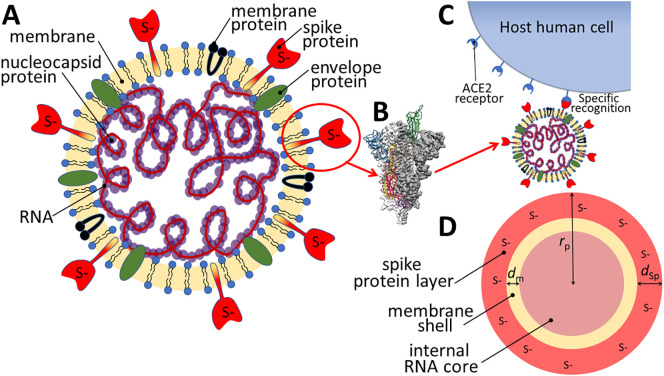
Fig. 2.
Schematic representation of (A) the SARS-CoV-2 virion, (B) a side view of the prefusion structure of spike protein with a single Receptor Binding Domain (RBD) in open (“up”) conformation (green), adapted from Wrapp et al. 2020 [4]. (C) ACE2-virion specific recognition, and (D) virion structure according to a physical-chemical relevant multilayered core-shell representation. Herein, the surface layer containing the spike protein is considered as the outer “shell” of the virion with thickness dSp (≈10 nm) hosting reactive sites S (corresponding to functional groups on the amino acid residues which may carry a negative, positive, or no net charge), the virion further comprises an intermediate membrane layer (thickness dm) and an internal RNA “core” with radius a = rp- dm- dSP (≈20 to 60 nm). In (A) and (D) the reactive sites S in the outer shell layer are nominally shown as negatively charged.