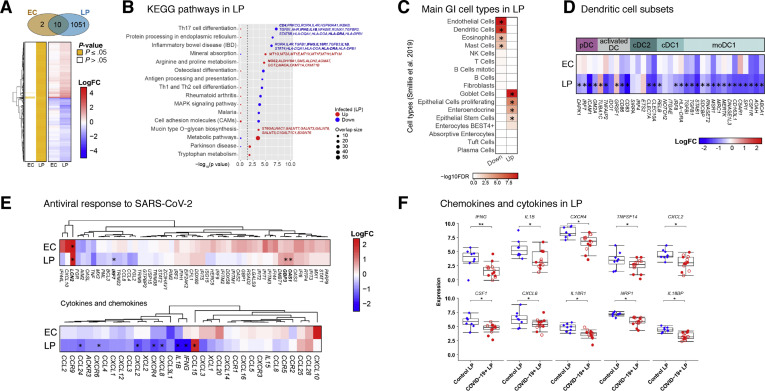
Figure 4.
Transcriptional changes in intestinal biopsy samples from patients with COVID-19 compared with control individuals. (A) Hierarchical clustering of average expression changes for 1063 genes (rows) with induced (red) or depleted (blue) expression (FDR, ≤0.05) in the EC and LP of intestinal biopsy samples from patients with COVID-19. The panel on the left indicates significant genes for each tissue fraction in yellow. The color bar indicates the average log2 fold change (FC). (B) The top enriched pathways (KEGG) that are induced (red) or depleted (blue) in the LP of patients with COVID-19 are displayed. The dashed line indicates the P ≤ .05 cutoff. Gene names are indicated for main pathways. (C) Deconvolution of main gastrointestinal cell types enriched or depleted in the LP of patients with COVID-19 compared with control individuals. Reference single cell RNA–seq cell-type signatures were taken from Smillie et al18 (P ≤ .05, Fisher exact test). (D) Average expression changes for DC markers in the EC and LP. Reference small conditional RNA–seq cell-type signatures were taken from Martin et al. The color bar indicates the average log2(FC). (E) Hierarchical clustering of average expression changes (columns) in the EC and LP for genes related to antiviral response to SARS-CoV-2 in post mortem lung tissue of patients with COVID-19, as described by Blanco-Mello et al (top) and for cytokines and chemokines (bottom). The color bar indicates the average log2 FC. (F) The gene expression levels for the top 10 significant chemokines and cytokines in the LP of patients with COVID-19 and control individuals. ∗P < .05, ∗∗P < .01. moDC, monocyte-derived dendritic cell.