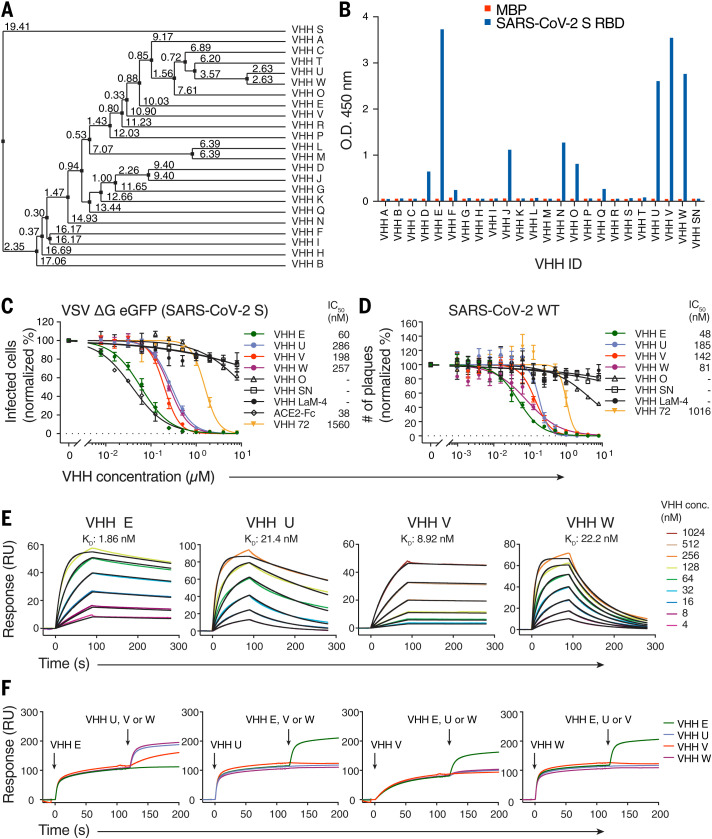
Fig. 1. Camelid nanobodies against two epitopes on the SARS-CoV-2 spike RBD neutralize infection.
(A) Average distance tree of nanobody candidates identified by phage display, calculated by percentage identity (66). (B) Binding of 100 nM HA-tagged VHHs to immobilized SARS-CoV-2 spike RBD or a control protein (MBP) was quantified by ELISA with horseradish peroxidase (HRP)–coupled anti-HA antibody. Unrelated VHH SN was used as a negative control. O.D., optical density. (C) SARS-CoV-2 spike–pseudotyped VSV ΔG eGFP was incubated with the indicated concentrations of HA- or LPETG-tagged (VHH LaM-4, VHH 72) VHHs or ACE2-Fc at 37°C for 1 hour, followed by infection of Vero E6 cells for 8 hours. eGFP-positive cells were measured by flow cytometry to quantify infection. Normalized values from three independent experiments ± SEM and IC50 values are plotted. (D) SARS-CoV-2 was incubated with the indicated concentrations of HA- or LPETG-tagged VHHs as in (C), followed by plaque assay on Vero E6 cells. Plaques were enumerated 3 days after infection; normalized values of three independent experiments ± SEM and IC50 values are plotted. (E and F) Biotinylated SARS-CoV-2 spike RBD was immobilized on SPR spectroscopy chips. (E) Indicated HA-tagged VHHs were injected for 90 s, followed by dissociation for 180 s. Dissociation constants (KD) were determined on the basis of fits, applying a 1:1 interaction model. (F) Epitope binning was performed by first injecting a single VHH for 120 s, followed by injection of a 1:1 mixture of the first nanobody in combination with VHH E, U, V, or W for 80 s.