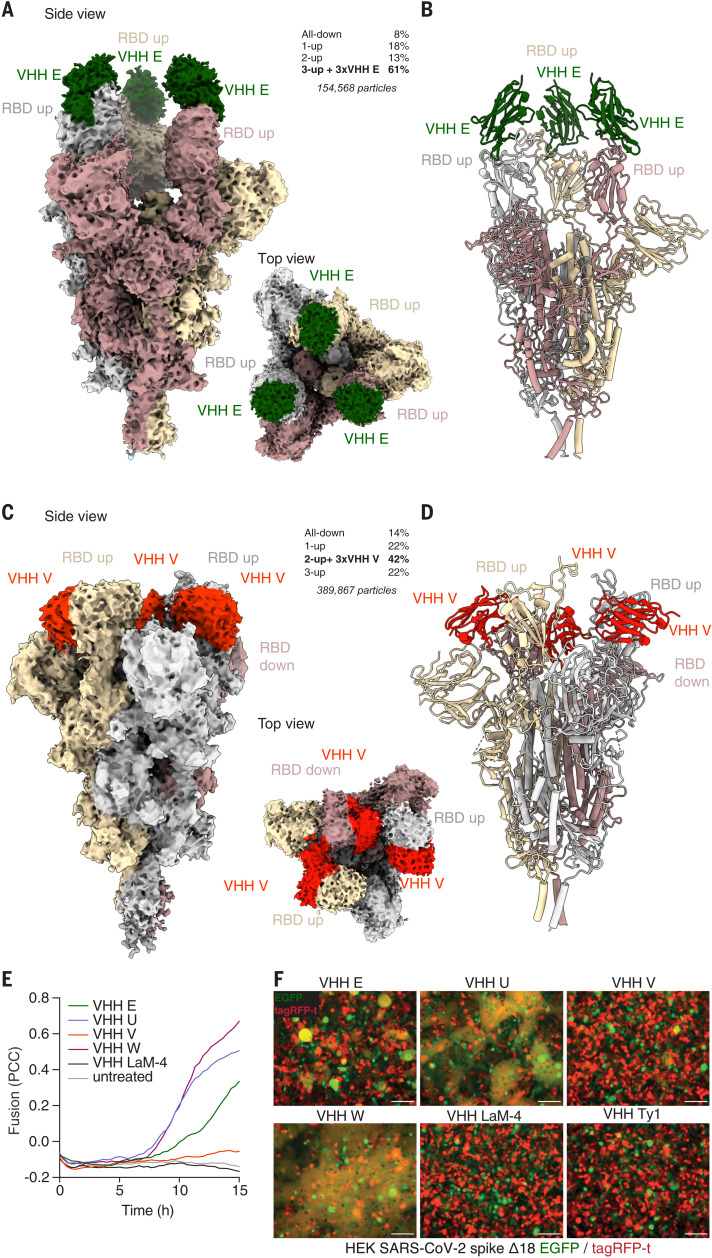
Fig. 3. Cryo-EM structures reveal that VHHs stabilize SARS-CoV-2 spike trimers with RBDs in the up conformation.
(A to D) Cryo-EM reconstructions [(A) and (C)] and atomic models [(B) and (D)] of VHH E [(A) and (B)] and VHH V [(C) and (D)] in complex with trimeric SARS-CoV-2 spike. Frequencies of the identified complexes as well as total numbers of considered particles are noted. (A and B) VHH E (in green) binds to SARS-CoV-2 in a 3-up conformation in the most abundant complex; the resolution is 3.3 Å [0.143 Fourier shell correlation (FSC)]. (C and D) VHH V (in red) binds to SARS-CoV-2 in a 2-up conformation with all VHH binding sites occupied at a resolution of 3.0 Å (0.143 FSC). In the most abundant complex, VHH V binds to the RBD in the up or the down conformation. (E and F) HEK 293 cells inducibly expressing SARS-CoV-2 S Δ18 and either eGFP or tagRFP-t were seeded into microscopy-grade 96-well plates in a 1:1 ratio and induced with 1 μg/ml doxycycline for 20 hours. Cells were treated with 1 μM of the indicated VHHs, and microscopy images were recorded every 20 min for 14 hours at 37°C. (E) Fusion was quantified by calculating Pearson correlation coefficients (PCC) between eGFP and tagRFP-t. Average values from four fields of view of an experiment representative of three independent experiments are displayed. (F) Representative images of cells 12 hours after treatment are displayed (also see fig. S13 and movies S8 to S13). Scale bars, 100 μm.