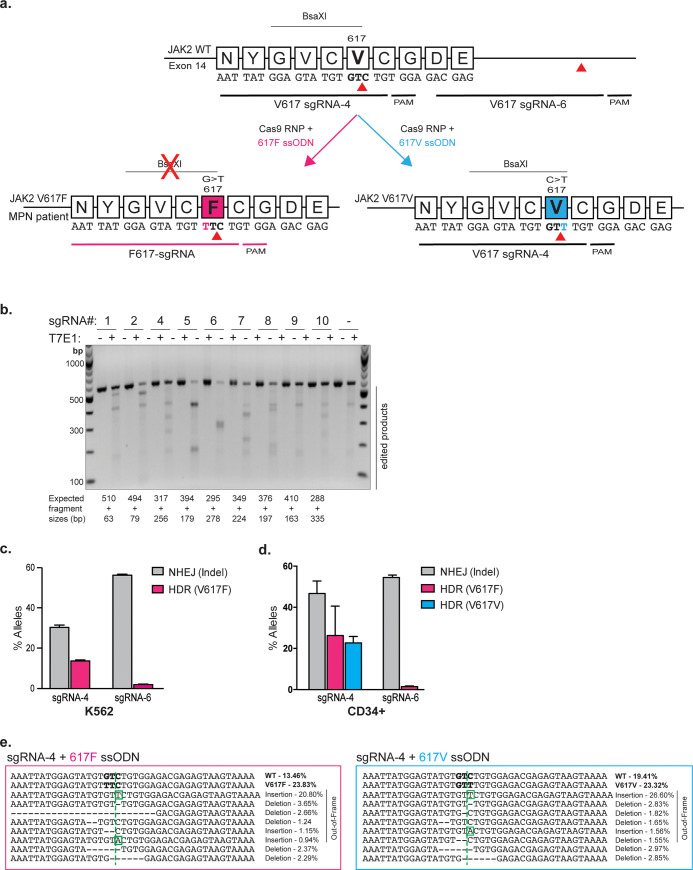
Fig 1. Cas9-RNP and ssODN-mediated targeting of the JAK2 locus in K562s and CD34+ HSPCs.
(a) Schematic of the JAK2 V617 locus with relevant genetic editing reagents annotated. SgRNAs-4 and -6 are identified with their corresponding 5’-NGG-3’ PAMs. Red arrowheads indicate cut site directed by these sgRNAs. The conversion of G>T results in the amino acid change from valine to phenylalanine (V617F, magenta) and removes the naturally occurring BsaXI restriction site. The conversion of C>T results in a silent mutation (V617V, blue) and retains the BsaXI restriction site. (b) T7 endonuclease I assay showing indel formation in cell pools edited independently with 9 different sgRNAs. (c) K562 cells were nucleofected with Cas9-RNP and the 617F ssODN. NHEJ- and HDR-mediated editing outcomes were assessed by amplicon-NGS. Data from n = 3 independent biological replicates with mean±SD graphed. (d) CD34+ HSPCs were nucleofected with Cas9-RNP and 617F or 617V ssODN. NHEJ- and HDR-mediated outcomes were assessed by amplicon-NGS. Data from n≥3 independent biological replicates with mean±SD graphed. (e) Allele spectra and corresponding percentages of alleles generated following editing with sgRNA-4 RNP and 617F or 617V ssODN in CD34+ HSPCs. Mutations causing frame-shifts are outlined.