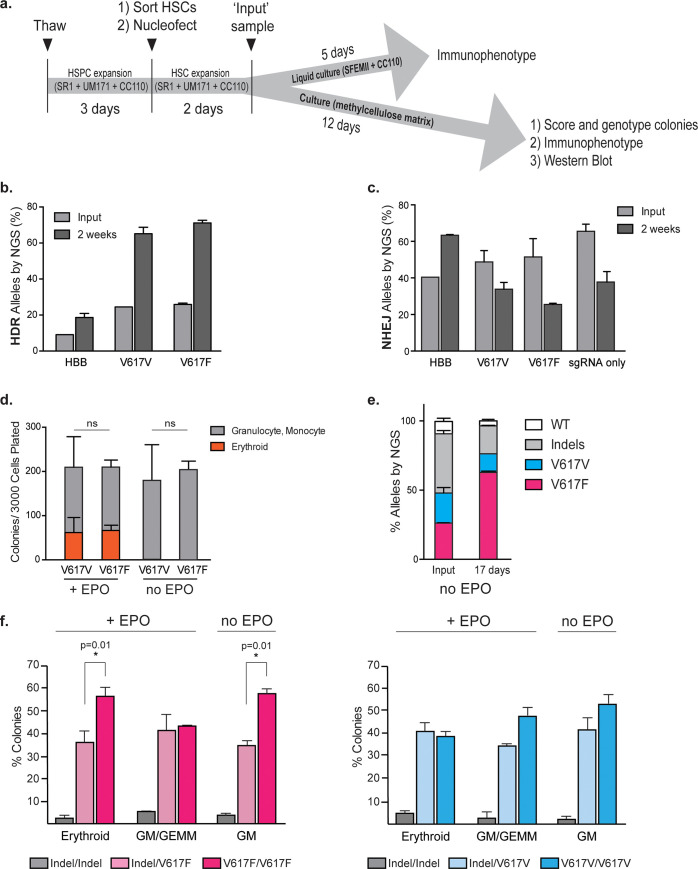
Fig 3. Identifying the enrichment of the V617F-edited alleles in CD34+ hematopoietic stem cells.
(a) Experimental layout for editing LT-HSCs. HSPCs were thawed and expanded for 3 days in SR1, UM171 and CC110. HSCs were sorted based on CD34+ CD38- CD45RA- CD90+ and nucleofected with appropriate editing reagents. Two days following electroporation an ‘input’ sample was taken to determine editing percentages. Cells were either maintained in liquid culture for immunophenotyping or plated in methylcellulose for further analyses. b. HSCs were nucleofected with Cas9-RNP and ssODNs encoding HBB, V617F or V617V mutations and were plated on methylcellulose as depicted in (a). HDR-mediated outcomes were assessed by NGS, two days (input) or two weeks after electroporation. Data from n = 3 independent biological replicates. Mean±SD shown. (c) NHEJ-mediated outcomes of cells in (b) were assessed by NGS, two days (input) or two weeks after electroporation. Data from n = 3 biological replicates. Mean±SD shown. (d) Pools of V167F- or V617V-edited HSCs were plated on methylcellulose with or without EPO and were scored as granulocyte, monocyte or erythroid based on morphology 14 days after plating. Data from n = 4 (for V617F) or n = 3 (for V617V) independent biological replicates. Mean±SD shown. (e) V617F- and V617V-edited HSCs were mixed and plated on methylcellulose. Input sample was taken directly after mixing prior to plating on methylcellulose. All resulting colonies were harvested two weeks later and processed for amplicon-NGS. Data from n = 4 biological replicates. Mean±SD shown. *:p<0.05 by unpaired t-test. (f) Single colonies were phenotyped as erythroid, GEMM, or GM and then genotyped by amplicon-NGS. V617F homozygous colonies were preferentially selected in the absence of EPO. Data from n≥3 biological replicates. Mean±SD shown. *:p<0.05 by unpaired t-test.