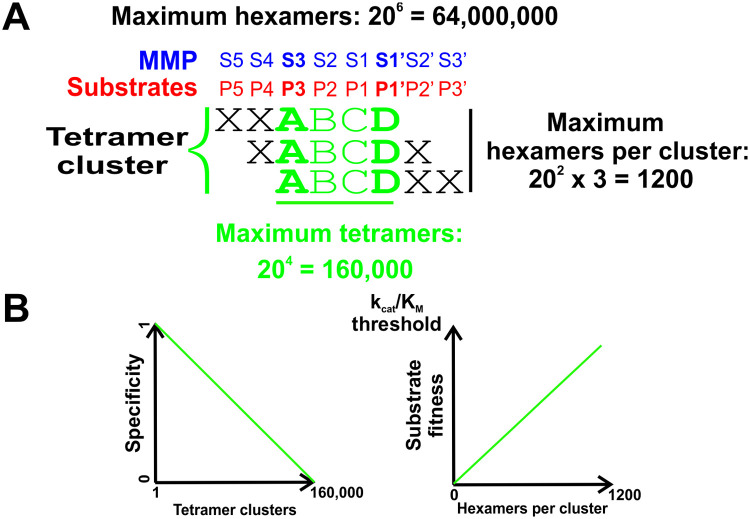
Fig 1. Basis for quantitative specificity profiling of the MMP catalytic cleft across S3 –S1՛.
A. Combinatorics of MMP–substrate interactions define the limits of substrate specificity and substrate fitness in experiments using hexapeptide probes. S3 and S1՛ are the most selective binding sites in the catalytic cleft of the MMPs (bold blue lettering). Together with the S2 and S1, they interact with P3-P1՛ tetramers in substrates (red lettering). To interrogate specificity of MMP-2 and 9, we used a library of randomized hexapeptides displayed on PIII gene product of M13 phage. The theoretical maximum of hexamer combinations is 64,000,000. The theoretical maximum for the number of hexapeptides containing identical tetramers (tetramer cluster) is 1200. There are 160,000 combinations of natural amino acid residues in random tetramers. B. Results of phage display analysis can be interpreted to quantify MMP specificity as well as the fitness of individual P3-P1՛ substrates. The number of tetramer clusters defines the amount of specificity of proteases recognizing P3-P1՛ positions in substrates, which ranges from absolutely specific (1 tetramer cluster) to absolutely non-specific (160,000 tetramer clusters). The number of hexamers per tetramer cluster is a measure of substrate fitness of all hexamers comprising it up to the kcat/KM threshold defined by experimental conditions.