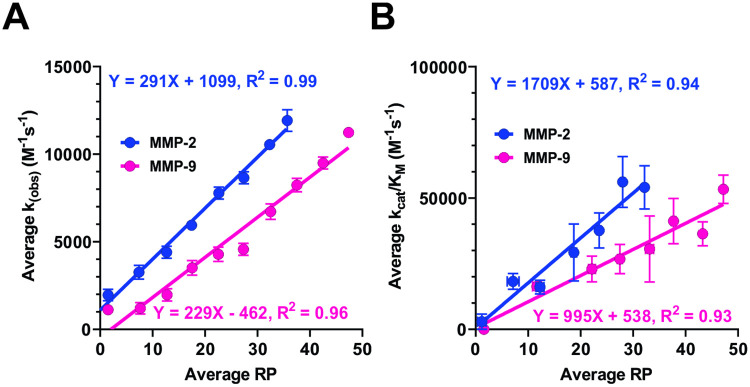
Fig 2. Probability of finding a tetramer cluster in substrate selections relative to the naïve library (RP) correlates with substrate fitness.
K(obs) values for 1369 individual phage substrates or kcat/KM values of 100 peptides derived from substrate phage sequences were experimentally determined as described in the text. The substrates were binned into evenly distributed groups based on the RP values of the tetramer clusters corresponding to their P3-P1՛ positions in substrates. The average K(obs) (A) or kcat/KM (B) values for each bin were plotted as a function of the corresponding average RP values and the data were subjected to linear regression analysis. The equations and goodness of fit parameters (R2) of the linear regression analyses of the MMP-2 and 9 data are shown at the top and bottom of the graph, respectively. These results can be compared to raw, unbinned data presented in the last graphs of the S6 Table, in the corresponding MMP specific tabs. The corresponding Pearson correlation coefficient R values for the raw (unbinned) data are 0.66 and 0.76 for MMP-2 and MMP-9, respectively, indicating already well correlated data.