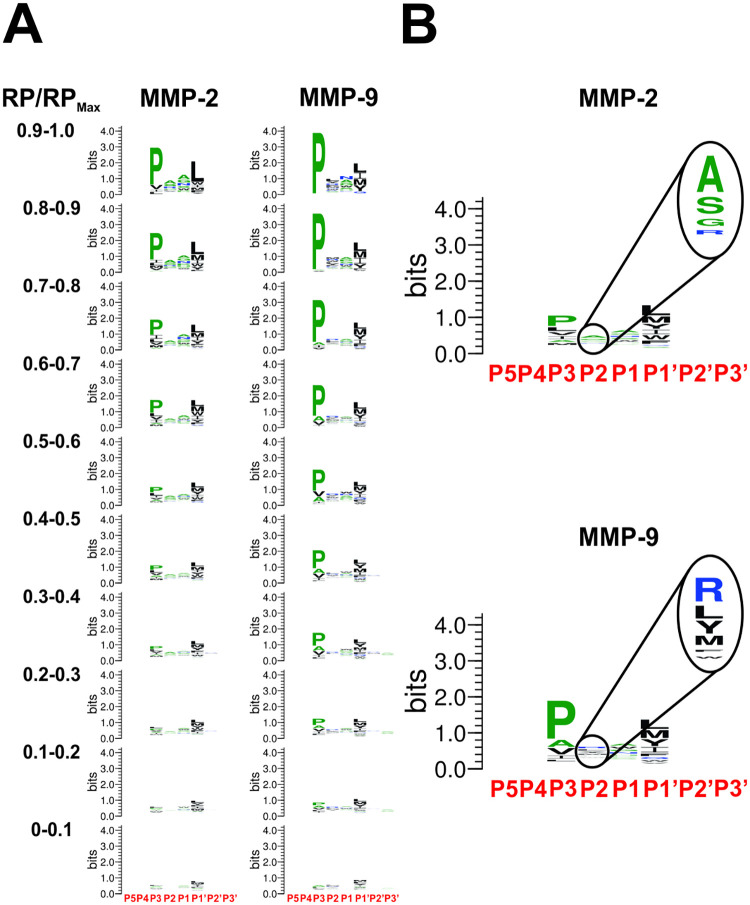
Fig 4. Composition of the selectomes of MMP-2 and 9 reflect differences in their specificities.
A. Selectome-based specificity profiles of MMP-2 and 9 reflect changes in substrate composition as a function of substrate fitness. Peptide hexamers in the selectomes of MMP-2 and 9 were aligned along the P5-P3՛ positions based on P3-P1՛ matches in the corresponding tetramer clusters and divided into 10 groups according to their RP values relative to the maximum (RP/RPMax). Relative abundances of amino acid residues at each position were calculated and presented in the form of a logo plot for each of the groups. B. Aggregate specificity profiles of MMP-2 and 9 reveal the major selectivity features of MMP-2 and 9. Hexamers belonging to the tetramer clusters constituting the selectomes of MMP-2 and 9 were aligned along the P5-P3՛ positions based on P3-P1՛ matches in the corresponding tetramer clusters, and relative abundances of residues at each position were plotted in the form of a logo. Zoom-in ovals show the relative contributions of residues to the P2 specificity profile of each enzyme.