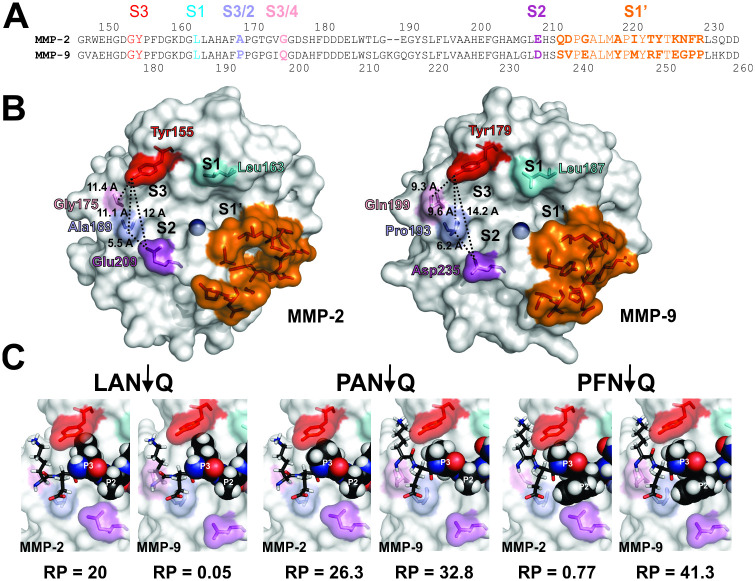
Fig 6. Changes in composition of selective substrates correlate with changes in selectivity determinants between MMP-2 and 9.
A. Distribution of the selectivity determinants across the subsites of the catalytic clefts of MMP-2 and 9. Sequences of the catalytic domains of MMP-2 and 9 were aligned based on the crystal structures of the catalytic domains of the respective enzymes (PDB IDs: 1QID for MMP2, 1GKC for MMP9). Residue numberings from the native N-termini are shown above (MMP-2) or below (MMP-9) the sequence. Selectivity Determining Positions (SDPs) are shown in color and marked in larger font. The catalytic cleft subsites they contribute to are shown directly above each SDP. Residues marked in bold differ between the two proteases (see text for details). B. Structural features of the SDPs in the catalytic clefts of MMP-2 and 9 provide basis for experimentally determined subsite selectivity. Residues contributing to the SDPs at S3, S2, S1 and S1՛ binding pockets are shown on the surface representations of the three-dimensional structures of MMP-2 and 9 in colors matching the sequence alignments in A. See text for more details. PyMOL molecular visualization system was used for display and analysis of 3D structures. C. Single substitutions in substrate tetramers illustrate distinctions in substrate recognition by MMP-2 and 9. Substrates selective for MMP-2 (KELAN↓Q), MMP-9 (KEPFN↓Q) and in common between the two enzymes (KEPAN↓Q) were docked into the catalytic clefts of MMP-2 and MMP-9 (See the Methods section for details). The P3-P1՛ sequences are shown above the structures. The docked residues below corresponding to the P3 and P2 residues in tetramers are shown as spheres, while the rest of the sequence is shown as sticks. Positions of residues relative to the scissile bond in the docked peptides are marked by white lettering. The RP values for the corresponding P3-P1՛ tetramer clusters are shown directly below the structures of each complex.