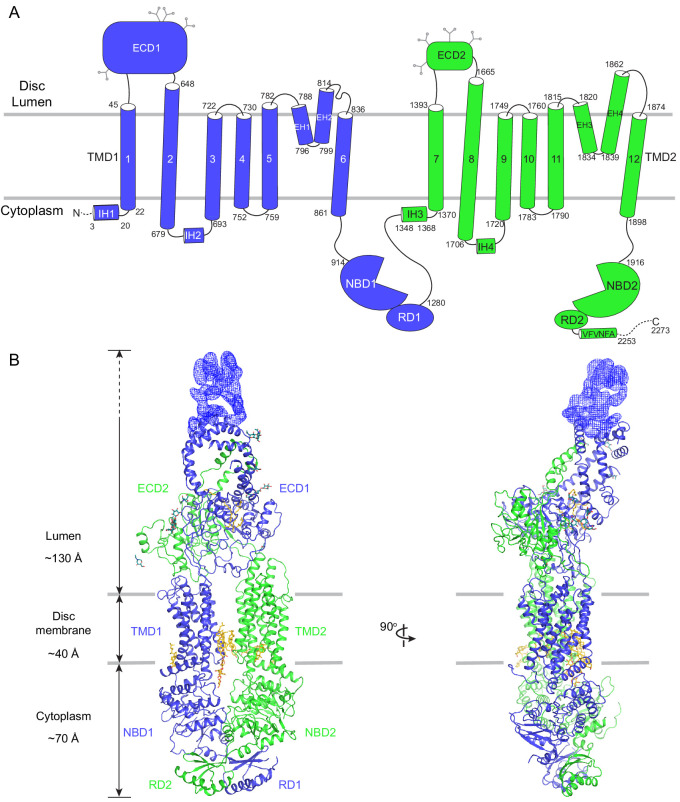
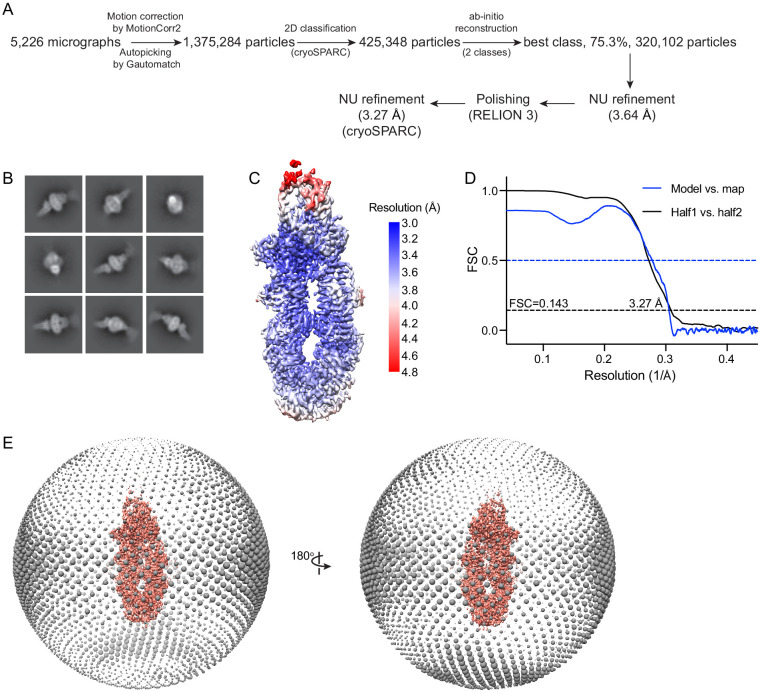
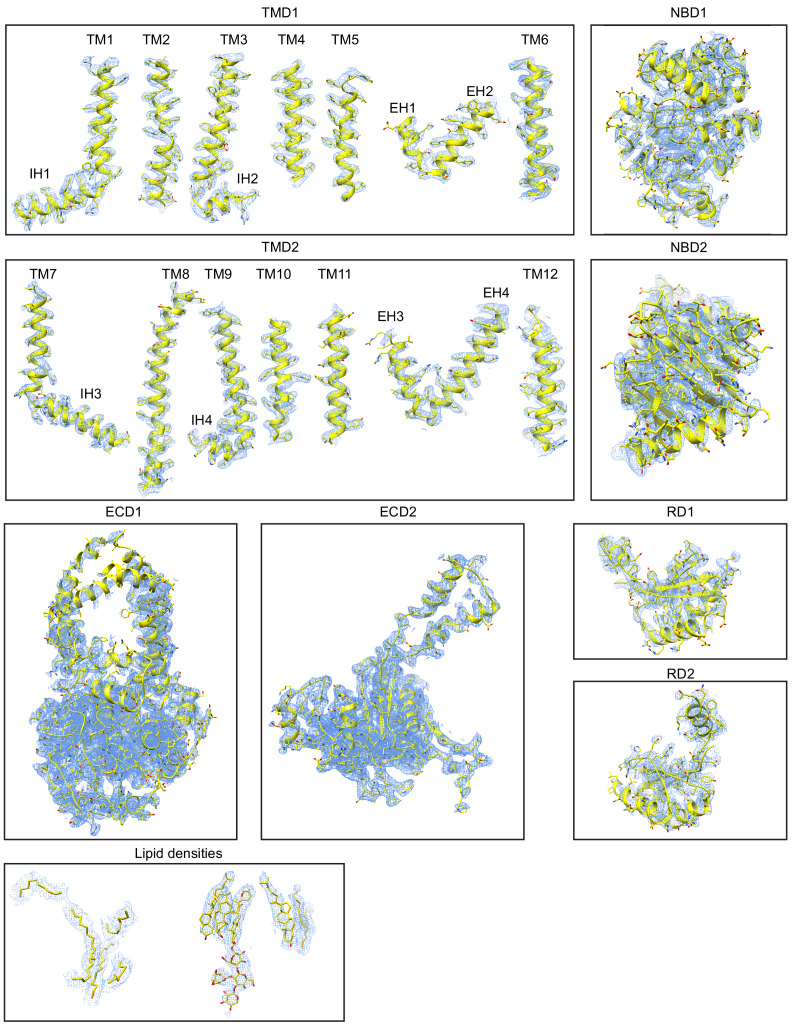
Figure 2. The overall structure of ABCA4 in the absence of ATP.
(A) The domain structure of ABCA4. The N- and C-terminal halves of the molecule are shown in blue and green. (B) Two orthogonal views of ABCA4 in ribbon presentation. Also shown is the electron microscopy density corresponding to residues 138–271 in ECD1. Ordered detergents, lipids, and N-linked glycans are shown as stick model. The position of the membrane is indicated by two gray lines.